Cloning and Functional Verification of Genes Related to 2-Phenylethanol Biosynthesis in Rosa rugosa
Abstract
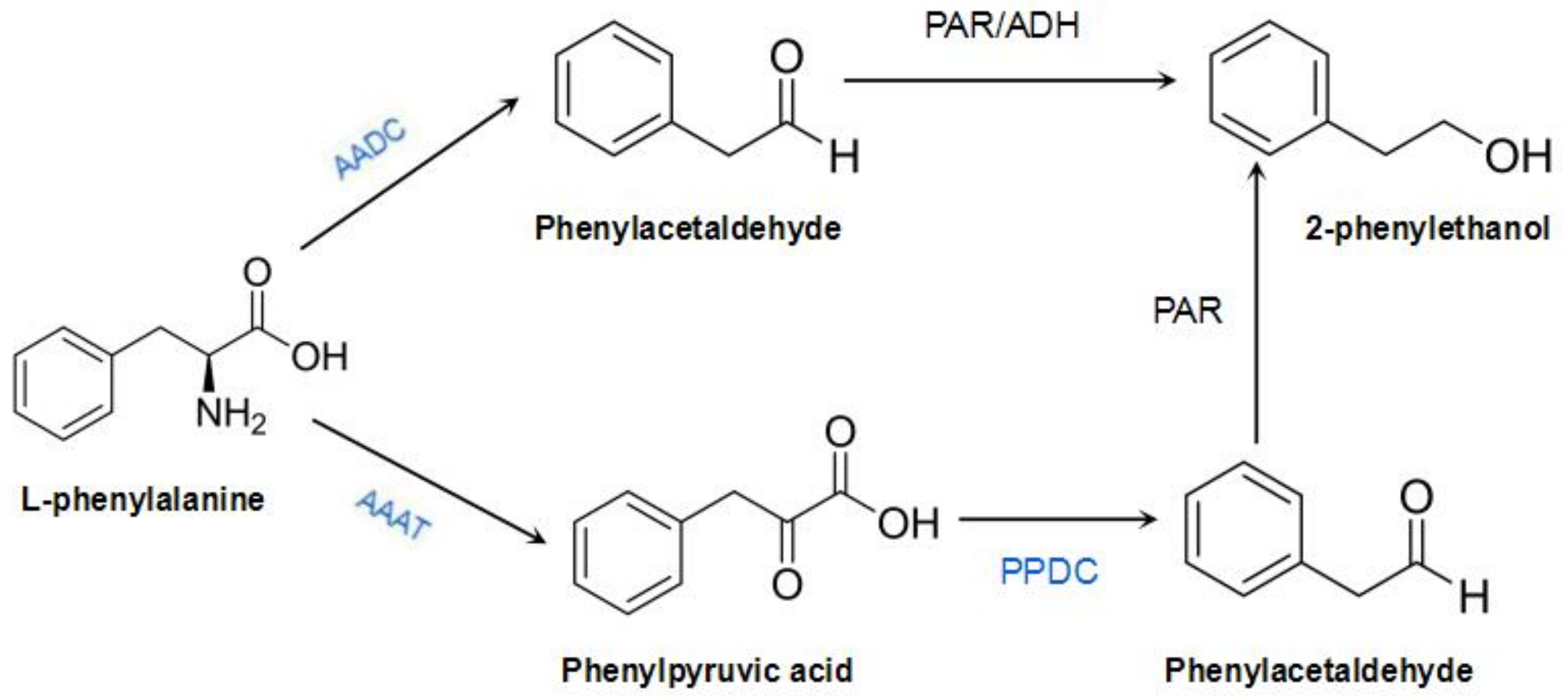
:1. Introduction
2. Materials and Methods
2.1. Cloning of the RrAAAT and RrPPDC1 Genes
2.1.1. RNA Extraction and Purification
2.1.2. 3′ Rapid-Amplification of cDNA Ends (RACE)
2.1.3. 5′ Rapid-Amplification of cDNA Ends
2.1.4. Purification, Cloning, and Sequencing
2.2. Sequence Analysis
2.3 Expression Analysis of RrAAAT and RrPPDC1 Genes in Rosa rugosa
2.4. Construction of the RrAADC, RrAAAT, and RrPPDC1 Overexpression Vectors
2.5. Plant Genetic Transformation and Phenotypic Analysis
2.6. Gas Chromatography with Mass Spectrometry Analysis of Floral Volatiles from Transgenic and Control Petunia Plants
3. Results
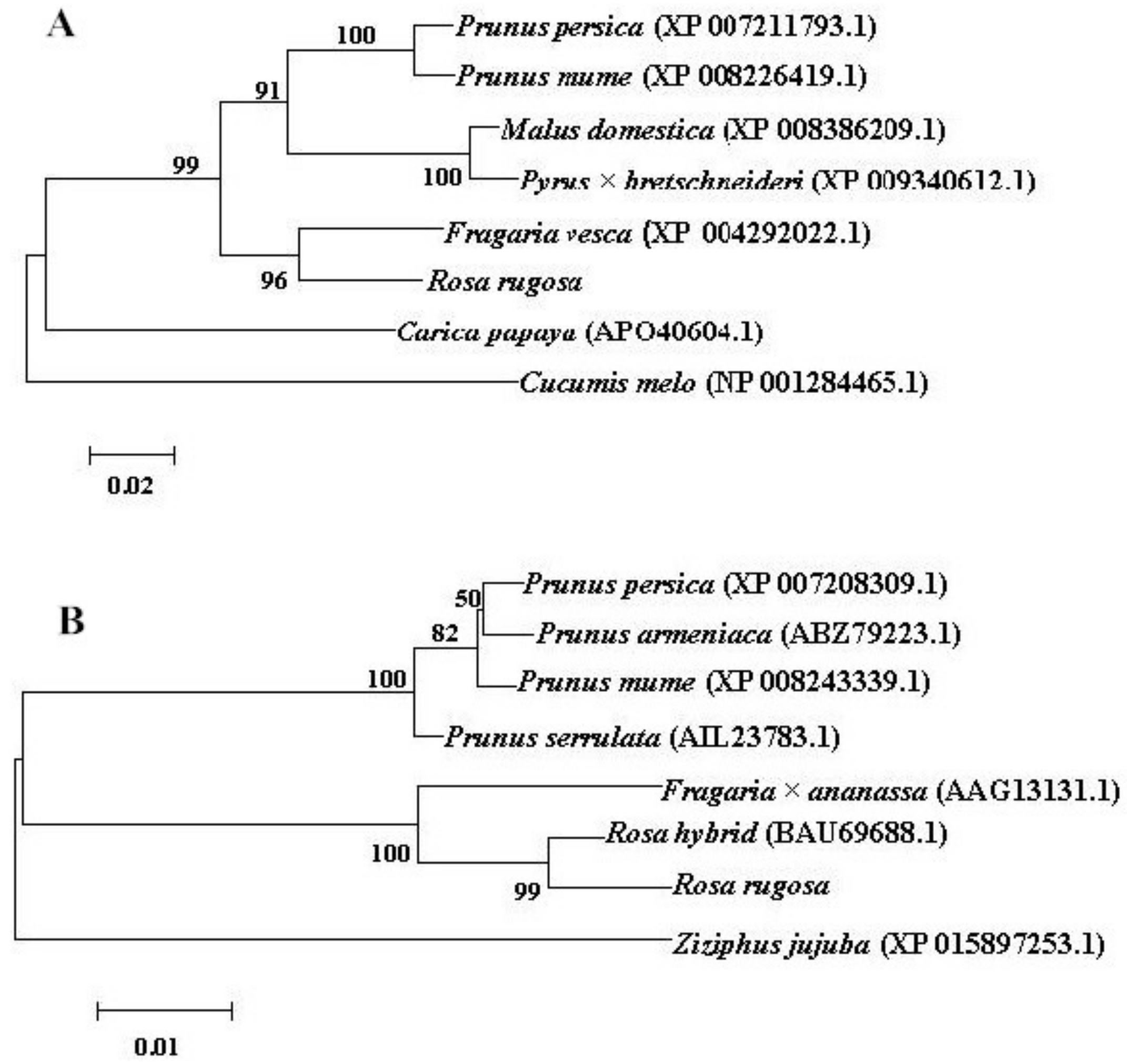
3.1. RrAAAT and RrPPDC1 Complementary DNA Isolation, Sequence Analysis, and Phylogenetic Tree
3.2. Temporal and Spatial Expression Analyses of RrAAAT and RrPPDC1 Genes in Rosa rugosa
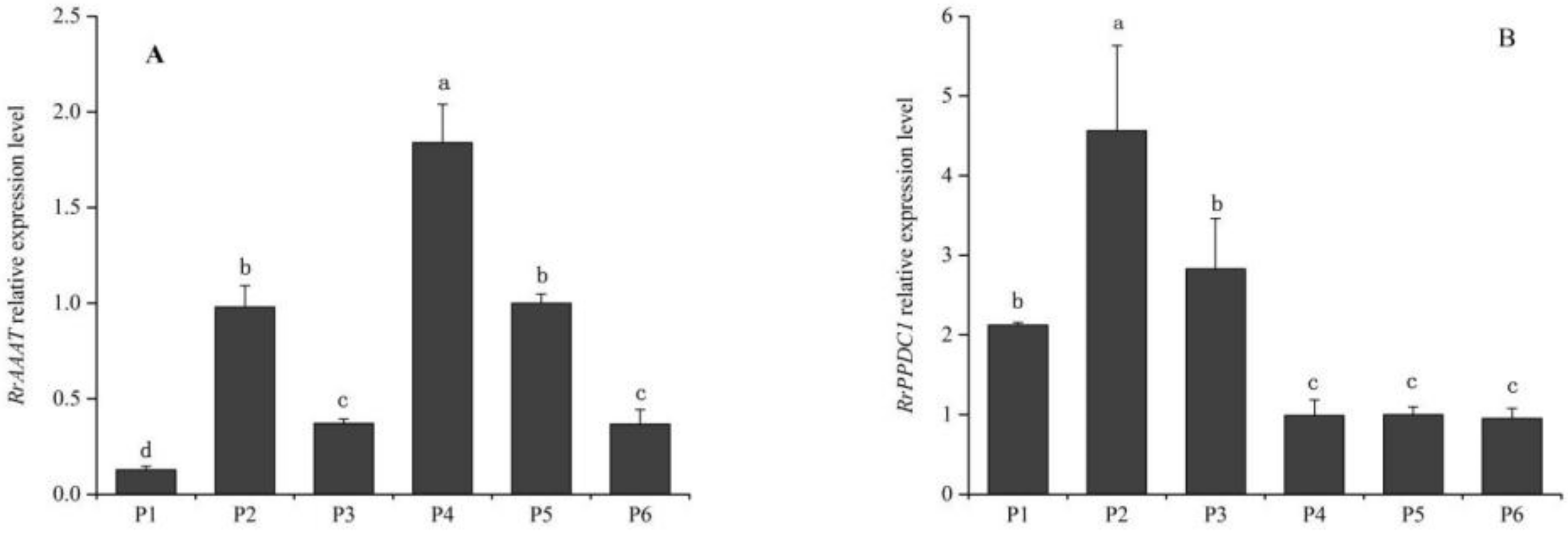
3.2.1. Analysis of RrAAAT and RrPPDC1 Expression Patterns in Rosa rugosa at Different Development Stages
3.2.2. Analysis of RrAAAT and RrPPDC1 Expression in Different Parts of Flower Organs in Rosa rugosa
3.3. Construction of the Overexpression Vectors, Plant Genetic Transformation, and Phenotypic Analysis
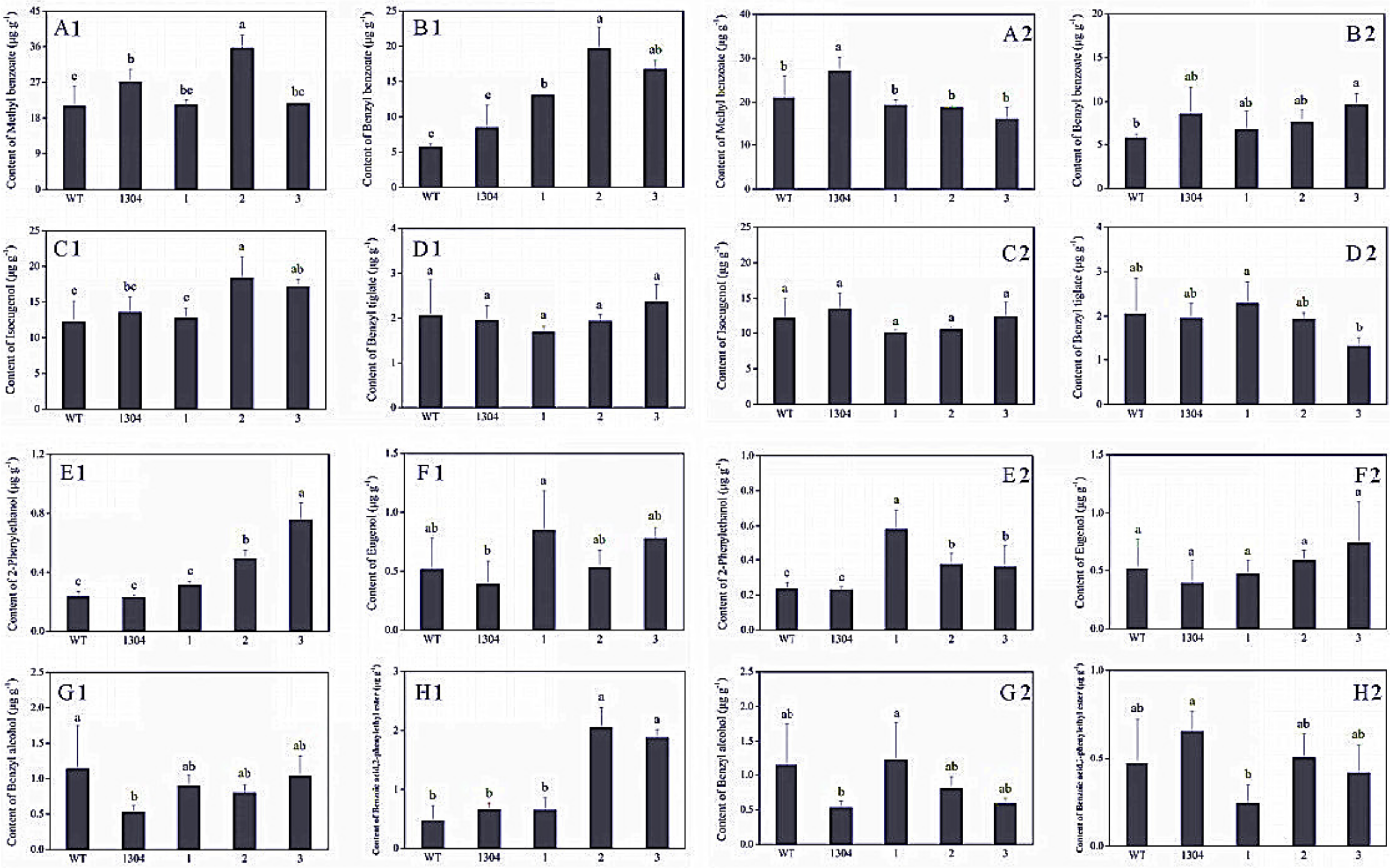
3.3.1. RrAADC and RrAAAT Overexpression Increases the Content of 2-Phenylethanol in Petunia
3.3.2. RrPPDC1 Overexpression Increases the Diameter of Petunia Flowers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Feng, L.G.; Chen, C.; Sheng, L.X.; Liu, P.; Tao, J.; Su, J.L.; Zhao, L.Y. Comparative analysis of headspace volatiles of Chinese Rosa rugosa. Molecules 2010, 15, 8390–8399. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.G.; Sheng, L.X.; Zhao, L.Y.; Yu, X.Y.; Shao, D.W.; He, X.D. Changes of the aroma constituents and contents in the course of Rosa rugosa Thunb. flower development. Sci. Agric. Sin. 2008, 41, 4341–4351. [Google Scholar]
- Yu, Y.L.; Wang, Q.Y.; Yao, L. Research on the aroma constituents and contents of Rosa rugosa ‘Purple Branch’. J. Shanghai Jiaotong Univ. (Agric. Sci.) 2012, 29, 80–87. [Google Scholar]
- Bugorskii, P.S.; Zaprometov, M.N. Biosynthesis of beta-phenylethanol in rose petals. Biokhimiia 1978, 43, 2038–2042. [Google Scholar] [PubMed]
- Hayashi, S.; Yagi, K.; Ishikawa, T.; Kawasaki, M.; Asai, T.; Picone, J.; Turnbull, C.; Hiratake, J.; Sakata, K.; Takada, M.; et al. Emission of 2-phenylethanol from its β-d-glucopyranoside and the biogenesis of these compounds from [2H8] l-phenylalanine in rose flowers. Tetrahedron 2004, 60, 7005–7013. [Google Scholar] [CrossRef]
- Watanabe, S.; Hayashi, K.; Yagi, K.; Asai, T.; MacTavish, H.; Picone, J.; Turnbull, C.; Watanabe, N. Biogenesis of 2-phenylethanol in rose flowers: Incorporation of [2H8] l-phenylalanine into 2-phenylethanol and its β-d-glucopyranoside during the flower opening of Rosa ‘Hoh-Jun’ and Rosa damascene Mill. Biosci. Biotechnol. Biochem. 2002, 66, 943–947. [Google Scholar] [CrossRef] [PubMed]
- Sakai, M.; Hirata, H.; Sayama, H.; Sekiguchi, K.; Itano, H.; Asai, T.; Dohra, H.; Hara, M.; Watanabe, N. Production of 2-phenylethanol in roses as the dominant floral scent compound from l-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci. Biotechnol. Biochem. 2007, 71, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Ohnishi, T.; Ishida, H.; Tomida, K.; Sakai, M.; Hara, M.; Watanabe, N. Functional characterization of aromatic amino acid aminotransferase involved in 2-phenylethanol biosynthesis in isolated rose petal protoplasts. J. Plant Physiol. 2012, 169, 444–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirata, H.; Ohnishi, T.; Watanabe, N. Biosynthesis of floral scent 2-phenylethanol in rose flowers. Biosci. Biotechnol. Biochem. 2016, 80, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Ohnishi, T.; Tomida, K.; Ishida, H.; Kanda, M.; Sakai, M.; Yoshimura, J.; Suzuki, H.; Ishikawa, T.; et al. Seasonal induction of alternative principal pathway for rose flower scent. Sci. Rep. 2016, 6, 20234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, L.G.; Wang, M.; Wang, J.; Zang, S.; Xia, W.; Sheng, L.X. Isolation of 2-phenylethanol biosynthesis related genes and their relationship with 2-phenylethanol accumulation in Rosa rugosa. Acta Physiol. Plant. 2015, 37, 256. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 36, 1101–1108. [Google Scholar] [CrossRef]
- Guo, Y.L.; Yu, Y.; Yang, Z.; Qin, X.T.; Ma, J.; Han, Y.; Yang, X.; Li, M.Y. Over-expressing PMADS20-SRDX repressor leads to the formation of ectopic trichome and stoma on petals and pistils in petunia. Acta Hortic. Sin. 2014, 41, 509–520. [Google Scholar]
- Mass Spectral Database for Windows; Standard Reference Data Program, U.S. Department of Commerce; NIST/EPA/NIH Mass Spectral Library (NIST 11); National Institute of Standards and Technology: Gaithersburg, MD, USA, 2012.
- Wiley Registry of Mass Spectral Data, 10th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013.
- Zhao, Y.Q.; Zhou, S.J.; Peng, P.H.; Pan, H.T.; Zhang, Q.X. Research advances in metabolic regulation and genetic engineering of floral scent. J. Trop. & Subtrop. Bot. 2011, 19, 381–390. [Google Scholar]
- Tieman, D.M.; Loucas, H.M.; Kim, J.Y.; Clark, D.G.; Klee, H.J. Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 2007, 68, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Kaminaga, Y.; Schnepp, J.; Peel, G.; Kish, C.M.; Ben-Nissan, G.; Weiss, D.; Orlova, I.; Lavie, O.; Rhodes, D.; Wood, K.; et al. Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 2006, 281, 23357–23366. [Google Scholar] [CrossRef] [PubMed]
- Gutensohn, M.; Klempien, A.; Kaminaga, Y.; Nagegowda, D.A.; Negre-Zakharov, F.; Huh, J.H.; Luo, H.; Weizbauer, R.; Mengiste, T.; Tholl, D.; et al. Role of aromatic aldehyde synthase in wounding/herbivory response and flower scent production in different Arabidopsis ecotypes. Plant. J. 2011, 66, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Taylor, M.; Schauer, N.; Fernie, A.R.; Hanson, A.D.; Klee, H.J. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 2006, 103, 8287–8292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, M.; Ovadia, R.; Perl, A.; Bar, E.; Lewinsohn, E.; Galili, G.; Oren-Shamir, M. Enhanced formation of aromatic amino acids increases fragrance without affecting flower longevity or pigmentation in Petunia × hybrida. Plant Biotechnol. J. 2015, 13, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Gonda, I.; Bar, E.; Portnoy, V.; Lev, S.; Burger, J.; Schaffer, A.A.; Tadmor, Y.; Gepstein, S.; Giovannoni, J.J.; Katzir, N.; et al. Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J. Exp. Bot. 2010, 61, 1111–1123. [Google Scholar] [CrossRef] [PubMed]



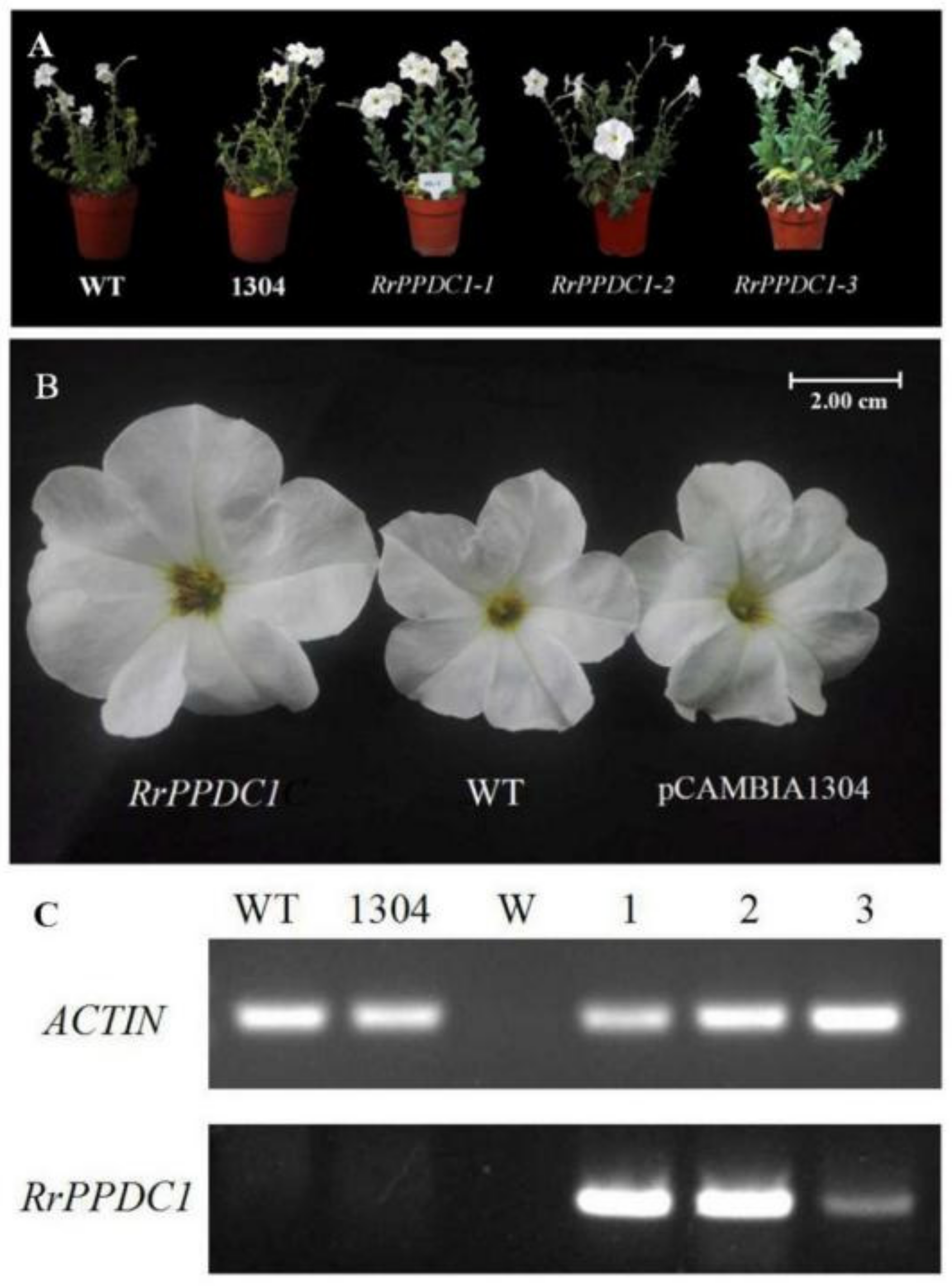

| WT | pCAMBIA1304 | RrPPDC1-1 | RrPPDC1-2 | RrPPDC1-3 |
|---|---|---|---|---|
| 49.43 ± 2.29 c | 49.00 ± 2.60 c | 56.14 ± 1.14 a | 53.56 ± 0.47 b | 55.34 ± 1.75 ab |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, L.; Zeng, Y.; Wei, T.; Zhu, M.; Fang, X.; Yuan, X.; Luo, Y.; Feng, L. Cloning and Functional Verification of Genes Related to 2-Phenylethanol Biosynthesis in Rosa rugosa. Genes 2018, 9, 576. https://doi.org/10.3390/genes9120576
Sheng L, Zeng Y, Wei T, Zhu M, Fang X, Yuan X, Luo Y, Feng L. Cloning and Functional Verification of Genes Related to 2-Phenylethanol Biosynthesis in Rosa rugosa. Genes. 2018; 9(12):576. https://doi.org/10.3390/genes9120576
Chicago/Turabian StyleSheng, Lixia, Yuqian Zeng, Tiantian Wei, Min Zhu, Xuemin Fang, Xiaoyu Yuan, Yunjian Luo, and Liguo Feng. 2018. "Cloning and Functional Verification of Genes Related to 2-Phenylethanol Biosynthesis in Rosa rugosa" Genes 9, no. 12: 576. https://doi.org/10.3390/genes9120576




