Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics
Abstract
1. Introduction
2. Cell-Free Protein Synthesis
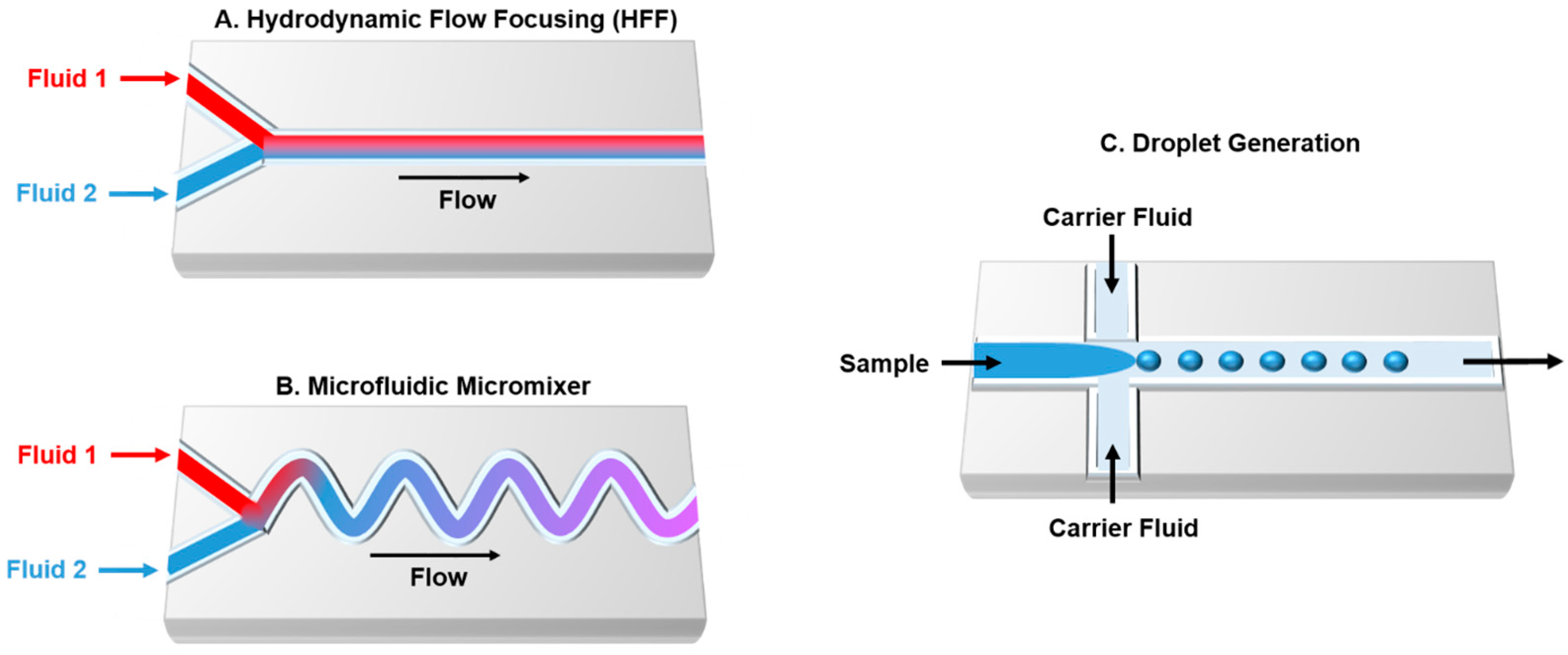
3. Microfluidic Technology
4. Microfluidic Platforms and Cell-Free Applications
4.1. De Novo Gene Synthesis in Microfluidics
4.2. Microfluidic Cell-Free Protein Synthesis
4.3. Microfluidics and Proteomics
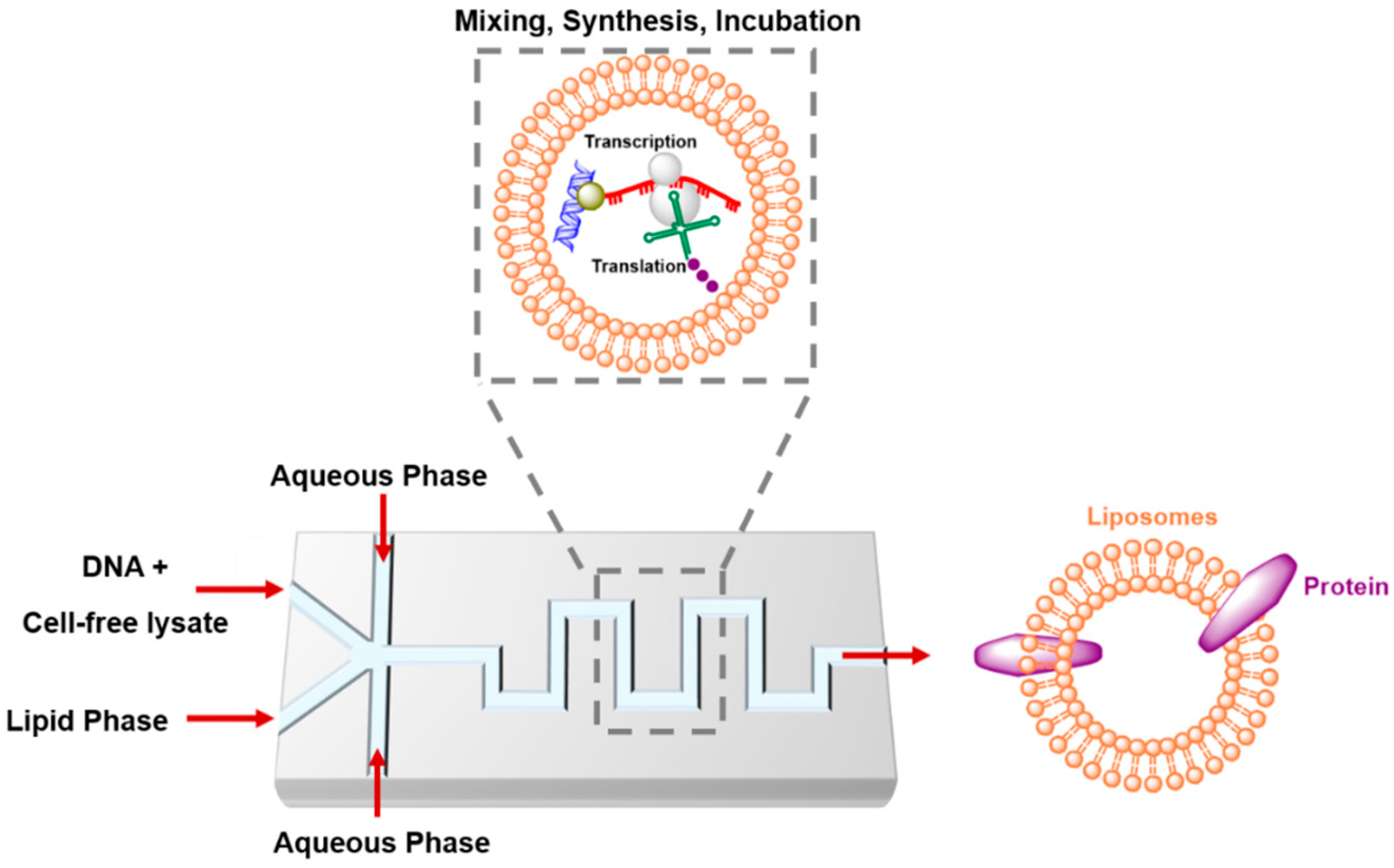
4.4. Artificial Cells in Microfluidics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hodgman, C.E.; Jewett, M.C. Cell-free synthetic biology: Thinking outside the cell. Metab. Eng. 2012, 14, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Smolke, C.D.; Silver, P.A. Informing biological design by integration of systems and synthetic biology. Cell 2011, 144, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Purnick, P.; Weiss, R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y. Cell-free synthetic biology: Engineering in an open world. Synth. Syst. Biotechnol. 2017, 2, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Linshiz, G.; Jensen, E.; Stawski, N.; Bi, C.; Elsbree, N.; Jiao, H.; Kim, J.; Mathies, R.; Keasling, J.D.; Hillson, N.J. End-to-end automated microfluidic platform for synthetic biology: From design to functional analysis. J. Biol. Eng. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Kou, S.; Cheng, D.; Sun, F.; Hsing, I.M. Microfluidics and microbial engineering. Lab Chip 2016, 16, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Gojobori, T. Marine metagenomics as a source for bioprospecting. Mar. Genom. 2015, 24, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Gojobori, T. Single-cell technologies in environmental omics. Gene 2016, 576, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.; Boutell, J.; Cooley, N.; He, M. Cell-free protein synthesis for proteomics. Brief. Funct. Genom. Proteom. 2004, 2, 308–319. [Google Scholar] [CrossRef]
- Stevens, R.C. Design of high throughput methods of protein production for structural biology. Struct. Fold. Des. 2000, 8, R177–R185. [Google Scholar] [CrossRef]
- Tymms, M.J. In vitro transcription and translation protocols’. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1995; Volume 37, pp. 1–4212. [Google Scholar]
- Endo, Y.; Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006, 17, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.; Gan, R.; Hodgman, E.; Jewett, M. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.V.; Miller, E.S. RNA binding properties of in vitro expressed histidine-tagged RB69 RegA translational repressor protein. Anal. Biochem. 1999, 269, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Swartz, J. Developing cell-free biology for industrial applications. J. Ind. Microbiol. Biotechnol. 2006, 33, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Merk, H.; Stiege, W.; Tsumoto, K. Cell-free expression of two singlechain monoclonal antibodies against lysozyme: Effect of domain arrangement on the expression. J. Biochem. 1999, 125, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Burks, E.A.; Chen, G.; Georgiou, G.; Iverson, B.L. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc. Natl. Acad. Sci. USA 1997, 94, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Inoue, A.; Tomari, Y. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, H.; Kanamori, T.; Shimizu, Y.; Ueda, T. A highly controllable reconstituted cell-free system—A breakthrough in protein synthesis research. Curr. Pharm. Biotechnol. 2010, 11, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Spirin, A.S.; Baranov, V.I.; Ryabova, L.A.; Ovodov, S.Y.; Alakhov, Y.B. A continuous cell-free translation system capable of producing polypeptides in high yield. Science 1988, 242, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Ramachandiran, V.; Kramer, G.; Hardesty, B. Expression of different coding sequences in cell-free bacterial and eukaryotic systems indicates translational pausing on E. coli ribosome. FEBS Lett. 2000, 482, 185–188. [Google Scholar] [CrossRef]
- Netzer, W.J.; Hartl, F.U. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 1997, 388, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, T.; Hasegawa, Y.; Tsuchimochi, M.; Kamura, N.; Ogasawara, T.; Kuroita, T.; Endo, Y. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 2002, 514, 102–105. [Google Scholar] [CrossRef]
- Kigawa, T.; Yabuki, T.; Yoshida, Y.; Tsutsui, M.; Ito, Y.; Shibata, T.; Yokoyama, S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999, 442, 15–19. [Google Scholar] [CrossRef]
- Matsuura, T.; Tanimura, N.; Hosoda, K.; Yomo, T.; Shimizu, Y. Reaction dynamics analysis of a reconstituted Escherichia coli protein translation system by computational modeling. Proc. Natl. Acad. Sci. USA 2016, 114, E1336–E1344. [Google Scholar] [CrossRef] [PubMed]
- Reyes, S.G.; Kuruma, Y.; Tsuda, S. Uncovering cell-free protein expression dynamics by a promoter library with diverse strengths. bioRxiv 2017. [Google Scholar] [CrossRef]
- Swartz, J.R. Transforming biochemical engineering with cell-free biology. AIChE J. 2012, 58, 5–13. [Google Scholar] [CrossRef]
- Kim, T.W.; Kim, D.M.; Choi, C.Y. Rapid production of milligram quantities of proteins in a batch cell-free protein synthesis system. J. Biotechnol. 2006, 124, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Swartz, J.R. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol. Bioeng. 1999, 66, 180–188. [Google Scholar] [CrossRef]
- Kim, D.M.; Choi, C.Y. A semicontinuous prokaryotic coupled transcription/translation system using a dialysis membrane. Biotechnol. Prog. 1996, 12, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Quast, R.B.; Sachse, R.; Schulze, C.; Wüstenhagen, D.A.; Kubick, S. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS ONE 2014, 9, e96635. [Google Scholar] [CrossRef] [PubMed]
- Kigawa, T.; Matsuda, T.; Yabuki, T.; Yokoyama, S. Bacterial cell-free system for highly efficient protein synthesis. In Cell-Free Protein Synthesis; Spirin, A.S., Swartz, J.R., Eds.; Wiley-VCH: Weinheim, Germany, 2008; pp. 83–97. [Google Scholar]
- Martin, G.A.; Kawaguchi, R.; Lam, Y.; DeGiovanni, A.; Fukushima, M.; Mutter, W. High-yield, in vitro protein expression using a continuous-exchange, coupled transcription/translation system. BioTechniques 2001, 31, 948–953. [Google Scholar] [PubMed]
- Takanori, K.; Takashi, Y.; Yasuhiko, Y.; Michio, T.; Yutaka, I.; Takehiko, S.; Shigeyuki, Y. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Letters 1999, 442. [Google Scholar] [CrossRef]
- Damiati, S.; Zayni, S.; Schrems, A.; Kiene, E.; Sleytr, U.; Chopineau, J.; Schuster, B.; Sinner, E.-K. Inspired and stabilized by nature: Ribosomal synthesis of the human voltage gated ion channel (VDAC) into 2D-protein-tethered lipid interfaces. Biomater. Sci. 2015, 3, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Robelek, R.; Lemker, E.S.; Wiltschi, B.; Kirste, V.; Naumann, R.; Oesterhelt, D.; Sinner, E.K. Incorporation of in vitro synthesized GPCR into a tethered artificial lipid membrane system. Angew. Chem. Int. Ed. Engl. 2007, 46, 605–608. [Google Scholar] [CrossRef] [PubMed]
- May, S.; Andreasson-Ochsner, M.; Fu, Z.K.; Low, Y.X.; Tan, D.; de Hoog, H.P.M.; Ritz, S.; Nallani, M.; Sinner, E.K. In Vitro Expressed GPCR Inserted in Polymersome Membranes for Ligand-Binding Studies. Angew. Chem. Int. Ed. Engl. 2013, 52, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, A.A.; Knoll, W.; Gennis, R.B.; Sinner, E.K. Cell-free synthesis of cytochrome bo(3) ubiquinol oxidase in artificial membranes. Anal. Biochem. 2012, 423, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Drese, K.S.; Latta, D.; Murr, A.; Ritzi-Lehnert, M.; Sinner, E.K. System for the In Vitro Transcription and Translation of Membrane Proteins. Patent WO 2011131231 A1, 27 October 2011. [Google Scholar]
- Damiati, S.; Kompella, U.B.; Damiati, S.A.; Kodzius, R. Microfluidic Devices for Drug Delivery Systems and Drug Screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Schneider, P.; Schneider, G. Accessing New Chemical Entities through Microfluidic Systems. Angew. Chem. Int. Ed. 2014, 53, 5750–5758. [Google Scholar] [CrossRef] [PubMed]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar] [CrossRef]
- Dittrich, P.S.; Manz, A. Lab-on-a-chip: Microfluidics in drug discovery. Nat. Rev. Drug Discov. 2006, 5, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Streets, A.; Huang, Y. Microfluidics for biological measurements with single-molecule resolution. Curr. Opin. Biotechnol. 2014, 25, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Gach, P.C.; Iwai, K.; Kim, P.W.; Hillson, N.J.; Singh, A.K. Droplet microfluidics for synthetic biology. Lab Chip 2017, 17, 3388–3400. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Castro, D.; Sumanpreet, K.C.; Parameswaran, A.M.; Foulds, I.G. DNA & Protein detection based on microbead agglutination. In Proceedings of the Hilton Head Workshop on the Science and Technology of Solid-State Sensors, Actuators, and Microsystems, Hilton Head, SC, USA, 3–7 June 2012. [Google Scholar]
- Kodzius, R.; Castro, D.; Foulds, I.G. Towards a high throughput droplet-based agglutination assay. In Proceedings of the 7th International Conference on Microtechnologies in Medicine and Biology (MMB), Marina del Rey, CA, USA, 10–12 April 2013. [Google Scholar]
- Huebner, A.; Sharma, S.; Srisa-Art, M.; Hollfelder, F.; Edel, J.B.; deMello, A.J. Microdroplets: A sea of applications? Lab Chip 2008, 8, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Lee, A. Microfluidic droplet manipulations and their applications. In Microdroplet Technology: Principles and Emerging Applications in Biology and Chemistry; Integrated Analytical Systems; Day, P., Manz, A., Zhang, Y., Eds.; Springer: New York, NY, USA, 2012; pp. 23–50. [Google Scholar]
- Wirth, T. Microreactors in Organic Synthesis and Catalysis; Wiley-VCH: Weinheim, Germany, 2008; pp. 1–41. [Google Scholar]
- Kodzius, R.; Xiao, K.; Wu, J.; Yi, X.; Gong, X.; Foulds, I.G.; Wen, W. Inhibitory effect of common microfluidic materials on PCR outcome. Sens. Actuators B Chem. 2012, 161, 349–358. [Google Scholar] [CrossRef]
- Unger, M.A.; Chou, H.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 1999, 288, 113–116. [Google Scholar] [CrossRef]
- Liu, J.; Enzelberger, M.; Quake, S.R. A nanoliter rotary device for polymerase chain reaction. Electrophoresis 2002, 23, 1531–1536. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Angew. Chem. 1998, 37, 550–575. [Google Scholar] [CrossRef]
- Vinuselvi, P.; Park, S.; Kim, M.; Park, J.; Kim, T.; Lee, S. Microfluidic Technologies for Synthetic Biology. Int. J. Mol. Sci. 2011, 12, 3576–3593. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Mathies, R.A. Integrated microfluidic systems for high performance genetic analysis. Trends Biotechnol. 2009, 27, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Snyder, T.M.; Quake, S.R. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 2010, 38, 2514–2521. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, S.Y.; Ding, B.; Fei, C.Y.; Wan, W.; Hu, D.M.; Du, R.K.; Zhou, X.C.; Hong, J.; Liu, H.Y.; et al. Design and construction of small perturbation mutagenesis libraries for antibody affinity maturation using massive microchip-synthesized oligonucleotides. J. Biotechnol. 2015, 194, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Hu, S.; Wan, W.; Xu, M.; Du, R.; Zhao, W.; Gao, X.; Liu, J.; Liu, H.; Hong, J. Effective Optimization of Antibody Affinity by Phage Display Integrated with High-Throughput DNA Synthesis and Sequencing Technologies. PLoS ONE 2015, 10, e0129125. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.S.; Carr, P.A.; Chen, L.; Zhang, S.G.; Jacobson, J.M. Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 2007, 35, e61. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.D.; Ma, K.S.; Saaem, I. Advancing high-throughput gene synthesis technology. Mol. Biosyst. 2009, 5, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Fredlake, C.P.; Hert, D.G.; Kan, C.W.; Chiesl, T.N.; Root, B.E.; Forster, R.E.; Barron, A.E. Ultrafast DNA sequencing on a microchip by a hybrid separation mechanism that gives 600 bases in 6.5 min. Proc. Natl. Acad. Sci. USA 2008, 105, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Irimia, D.; Mindrinos, M.; Russom, A.; Xiao, W.Z.; Wilhelmy, J.; Wang, S.L.; Heath, J.D.; Kurn, N.; Tompkins, R.G.; Davis, R.W. Genome-wide transcriptome analysis of 150 cell samples. Integr. Biol. 2009, 1, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Au, L.C.; Yang, F.Y.; Yang, W.J.; Lo, S.H.; Kao, C.F. Gene synthesis by a LCR-based approach: High-level production of leptin-L54 using synthetic gene in Escherichia coli. Biochem. Biophys. Res. Commun. 1998, 248, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, W.; Crameri, A.; Ha, K.; Brennan, T.; Heyneker, H. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 1995, 164, 49–53. [Google Scholar] [CrossRef]
- Yehezkel, B.; Rival, A.; Raz, O.; Cohen, R.; Marx, Z.; Camara, M.; Dubern, J.F.; Koch, B.; Heeb, S.; Delattre, C.; et al. Synthesis and cell-free cloning of DNA libraries using programmable microfluidics. Nucleic Acids Res. 2016, 44, e35. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tangen, U.; Minero, G.A.; Sharma, A.; Wagler, P.F.; Cohen, R.; Raz, O.; Marx, T.; Ben-Yehezkel, T.; McCaskill, J.S. DNA-library assembly programmed by on-demand nano-liter droplets from a custom microfluidic chip. Biomicrofluidics 2015, 9, 044103. [Google Scholar] [CrossRef] [PubMed]
- Ochs, C.J.; Abate, A.R. Rapid modulation of droplet composition with pincer microvalves. Lab Chip 2015, 15, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Tewhey, R.; Warner, J.B.; Nakano, M.; Libby, B.; Medkova, M.; David, P.H.; Kotsopoulos, S.K.; Samuels, M.L.; Hutchison, J.B.; Larson, J.W. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009, 27, 1025–1031. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Paper-Based Synthetic Gene Networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Yi, X.; Xiao, K.; Li, S.; Kodzius, R.; Qin, J.; Wen, W. Wax-bonding 3D microfluidic chips. Lab Chip 2010, 10, 2622–2627. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Kodzius, R.; Gong, X.; Xiao, K.; Wen, W. A simple method of fabricating mask-free microfluidic devices for biological analysis. Biomicrofluidics 2010, 4, 036503. [Google Scholar] [CrossRef] [PubMed]
- Sawasaki, T.; Ogasawara, T.; Morishita, R.; Endo, Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 2002, 99, 14652–14657. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T. Beating the odd. Nat. Chem. Biol. 2016, 12, 54–55. [Google Scholar] [CrossRef] [PubMed]
- Khnouf, R.; Olivero, D.; Jin, S.; Fan, Z.H. Miniaturized fluid array for high-throughput protein expression. Biotechnol. Prog. 2010, 26, 1590–1596. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.; Maerkl, S.J.; Quake, S.R. An in vitro microfluidic approach to generating protein-interaction networks. Nat. Methods 2009, 6, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Kanter, G.; Yang, J.; Voloshin, A.; Levy, S.; Swartz, J.R.; Levy, R. Cell-free production of scFv fusion proteins: An efficient approach for personalized lymphoma vaccines. Blood 2007, 109, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Timm, A.; Shankles, P.; Foster, C.; Doktycz, M.; Retterer, S. Toward Microfluidic Reactors for Cell-Free Protein synthesis at the Point-of-Care. Small 2016, 12, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Hugh Fan, Z.; Mei, Q.; Khnouf, R.; Jin, S. Microfluidic protein synthesis array for toxin detection. In Proceedings of the Solid-State Sensors, Actuators and Microsystems Conference, (Transducers 2009), Denver, CO, USA, 21–25 June 2009. [Google Scholar]
- Maerkl, S.J. Next generation microfluidic platforms for high-throughput protein biochemistry. Curr. Opin. Biotechnol. 2011, 22, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ridgeway, W.K.; Seitaridou, E.; Phillips, R.; Williamson, J.R. RNA—Protein binding kinetics in an automated microfluidic reactor. Nucleic Acids Res. 2009, 37, e142. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kim, D.M. Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments. Biotechnol. J. 2013, 8, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Boozer, C.; Kim, G.; Cong, S.; Guan, H.; Londergan, T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: A review of new surface plasmon resonance technologies. Curr. Opin. Biotechnol. 2006, 17, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Joung, H.A.; Ahn, J.H.; Kim, K.O. Real-time monitoring of cell-free protein synthesis on a surface plasmon resonance chip. Anal. Biochem. 2007, 366, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Seefeld, T.H.; Halpern, A.R.; Corn, R.M. On-chip synthesis of protein microarrays from DNA microarrays via coupled in vitro transcription and translation for surface plasmon resonance imaging biosensor applications. J. Am. Chem. Soc. 2012, 134, 12358–12361. [Google Scholar] [CrossRef] [PubMed]
- Georgi, V.; Blechert, G.; Zwanzig, M.; Wüstenhagen, D.A.; Bier, F.; Jung, E.; Kubick, S. On-chip automation of cell-free protein synthesis: New opportunities due to a novel reaction mode. Lab Chip 2016, 16, 269–281. [Google Scholar] [CrossRef] [PubMed]
- He, M. Cell-free protein synthesis: Applications in proteomics and biotechnology. New Biotechnol. 2008, 25, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Du, W.; Luo, Q.; Liu, B.F. Microfluidic chip: Next-generation platform for systems biology. Anal. Chim. Acta 2009, 650, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.T.; Li, Y.; Woolley, A.T. Phase-Changing Sacrificial Materials for Interfacing Microfluidics with Ion-Permeable Membranes to Create On-Chip Preconcentrators and Electric Field Gradient Focusing Microchips. Anal. Chem. 2006, 78, 2565–2570. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Stevens, A.L.; Han, J. Million-fold Preconcentration of Proteins and Peptides by Nanofluidic Filter. Anal. Chem. 2005, 77, 4293–4299. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Burns, M.A.; Hasselbrink, E.F. Electrokinetic Protein Preconcentration Using a Simple Glass/Poly(dimethylsiloxane) Microfluidic Chip. Anal. Chem. 2006, 78, 4779–4785. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.J.; Fluri, K.; Seiler, K.; Fan, Z.; Effenhauser, C.S.; Manz, A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science 1993, 261, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Manz, A.; Miyahara, Y.; Miura, J.; Watanabe, Y.; Miyagi, H.; Sato, K. Design of an open-tubular column liquid chromatograph using silicon chip technology. Sens. Actuators B 1990, 1, 249–255. [Google Scholar] [CrossRef]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.A.; Whitesides, G.M. Rapid Prototyping of Microfluidic Systems in Poly-(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef] [PubMed]
- Michels, D.A.; Hu, S.; Schoenherr, R.M.; Eggertson, M.J.; Dovichi, N.J. Fully automated two-dimensional capillary electrophoresis for high sensitivity protein analysis. Mol. Cell. Proteom. 2002, 1, 69–74. [Google Scholar] [CrossRef]
- Li, Y.; Buch, J.S.; Rosenberger, F.; DeVoe, D.L.; Lee, C.S. Integration of Isoelectric Focusing with Parallel Sodium Dodecyl Sulfate Gel Electrophoresis for Multidimensional Protein Separations in a Plastic Microfluidic Network. Anal. Chem. 2004, 76, 742–748. [Google Scholar] [CrossRef] [PubMed]
- McClain, M.A.; Culbertson, C.T.; Jacobson, S.C.; Allbritton, N.L.; Sims, C.E.; Ramsey, J.M. Microfluidic Devices for the High-Throughput Chemical Analysis of Cells. Anal. Chem. 2003, 75, 5646–5655. [Google Scholar] [CrossRef] [PubMed]
- Ros, A.; Hellmich, W.; Regtmeier, J.; Duong, T.T.; Anselmetti, D. Bioanalysis in structured microfluidic systems. Electrophoresis 2006, 27, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Luisi, P.L.; Ferri, F.; Stano, P. Approaches to semi-synthetic minimal cells: A review. Naturwissenschaften 2006, 93, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stano, P.; Luisi, P.L. Semi-synthetic minimal cells: Origin and recent developments. Curr. Opin. Biotechnol 2013, 24, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing life. Nature 2001, 409, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Chuang, R.Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hu, S.; Chen, X. Artificial cells: From basic science to applications. Mater. Today 2016, 19, 516–532. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Noireaux, V. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 2012, 1, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Kuruma, Y.; Stano, P.; Ueda, T.; Luisi, P.L. A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim. Biophys. Acta 2009, 1788, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Tan, C. The engineering of artificial cellular nanosystems using synthetic biology approaches. Wiley Interdisciplin. Rev. Nanomed. Nanobiotechnol. 2014, 6, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Elani, Y.; Law, R.V.; Ces, O. Protein synthesis in artificial cells: Using compartmentalization for spatial organisation in vesicle bioreactors. Phys. Chem. Chem. Phys. 2015, 17, 15534–15537. [Google Scholar] [CrossRef] [PubMed]
- Karzbrun, E.; Tayar, A.M.; Noireaux, V.; Bar-Ziv, R.H. Programmable on-chip DNA compartments as artificial cells. Science 2014, 345, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.K.; Murray, V.L.; Liu, A.P. Engineering artificial cells by combining HeLa based cell-free expression and ultrathin double emulsion template. Methods Cell Biol. 2015, 128, 303–318. [Google Scholar] [PubMed]
- Matsubayashi, H.; Ueda, T. Purified cell-free systems as standard parts for synthetic biology. Curr. Opin. Biotechnol. 2014, 22, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Heymann, M.; Bernhard, F.; Schwille, P.; Kai, L. Cell-free protein synthesis in micro compartments: Building a minimal cell from biobricks. New Biotechnol. 2017, 39, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Elani, Y. Construction of membrane-bound artificial cells using microfluidics: A new frontier in bottom-up synthetic biology. Biochem. Soc. Trans. 2016, 44, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Teh, S.Y.; Lin, R.; Hung, L.H.; Lee, A.P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Rouilly, V.; Niu, X.; Chappell, J.; Kitney, R.I.; Edel, J.B.; Freemont, P.S.; deMello, A.J. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 2009, 6, S493–S506. [Google Scholar] [CrossRef] [PubMed]
- Courtois, F.; Olguin, L.F.; Whyte, G.; Bratton, D.; Huck, W.; Abell, C.; Hollfelder, F. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem 2008, 9, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Chanasakulniyom, M.; Martino, C.; Paterson, D.; Horsfall, L.; Rosser, S.; Cooper, J.M. Expression of membrane-associated proteins within single emulsion cell facsimiles. Analyst 2012, 137, 2939–2943. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ho, K.; Murray, V.; Liu, A. Engineering artificial cells by combining HeLa-based cell-free expression and ultrathin double emulsion template. In Methods in Cell Biology; Jennifer, R., Wallace, F.M., Eds.; Academic Press: New York, NY, USA, 2015; Volume 128, pp. 303–318. [Google Scholar]
- Kamiya, K.; Kawano, R.; Osaki, T.; Akiyoshi, K.; Takeuchi, S. Cell-sized asymmetric lipid vesicles facilitate the investigation of asymmetric membranes. Nat. Chem. 2016, 8, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, E.; Spruijt, E.; Hansen, M.; Dubuc, E.; Groen, J.; Chokkalingam, V.; Piruska, A.; Heus, H.A.; Huck, W. Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc. Natl. Acad. Sci. USA 2013, 110, 11692–11697. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]

| Cell-Based System | Versus | Cell-Free System |
|---|---|---|
| Yes | Needs cloning | No |
| No | Ability to produce toxic protein | Yes |
| No | Ability to express multiple genes | Yes |
| No | Usually generate functional, soluble, and folded proteins | Yes |
| No | Possible adjusting and controlling by the addition of helper molecules | Yes |
| No | Possible incorporation of non-natural or chemically modified amino acids | Yes |
| Yes | Native environment | No |
| Days | Time | Hours |
| Low | Costs | High |
| Escherichia coli Extract | Rabbit Reticulocyte Lysate | Wheat Germ Extract | Yeast Cells, Tumor Cells, Insects | |
|---|---|---|---|---|
| Protein yields | High (mg) | Low (µg) | High (mg) | Low (µg) |
| Generated proteins | Many incomplete polypeptides | Mainly full-length, folded proteins | Mainly full-length, folded, multidomain proteins | Mainly full-length, folded, multidomain proteins |
| Translation modifications | Post-translation | Co-translation | Co-translation | Co-translation |
| Recommended template sources | Bacteria | Prokaryotic (bacteria, mammalian virus, plant virus), Eukaryotic (plants, animals) | Prokaryotic (bacteria, plant virus), Eukaryotic (plants, animals) | - |
| Genetic modification tools | Well established | Poor | Poor | Poor |
| Extract preparation | Simple | Requires complex manipulation of animal tissues but cell breakage is easy and fast | Complex and time- consuming | Cell cultivation is complex and time-consuming, but cell breakage is easy and fast |
| Cost | Low | High | Low | High |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.-K. Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics. Genes 2018, 9, 144. https://doi.org/10.3390/genes9030144
Damiati S, Mhanna R, Kodzius R, Ehmoser E-K. Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics. Genes. 2018; 9(3):144. https://doi.org/10.3390/genes9030144
Chicago/Turabian StyleDamiati, Samar, Rami Mhanna, Rimantas Kodzius, and Eva-Kathrin Ehmoser. 2018. "Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics" Genes 9, no. 3: 144. https://doi.org/10.3390/genes9030144
APA StyleDamiati, S., Mhanna, R., Kodzius, R., & Ehmoser, E.-K. (2018). Cell-Free Approaches in Synthetic Biology Utilizing Microfluidics. Genes, 9(3), 144. https://doi.org/10.3390/genes9030144





