Abstract
The anaerobic membrane bioreactors (AnMBR) with ring membrane module were operated under mesophilic temperature (M-AnMBR) and atmospheric temperature (A-AnMBR). Compared to the M-AnMBR, the removal efficiency of the A-AnMBR was found to be lower and the faster membrane fouling occurred in the A-AnMBR under corresponding hydraulic retention time (HRT). The MiSeq high-throughput sequencing was applied to analyze the microbial community structure. The HRT change had different effects on the community richness and diversity of the cake and bulk sludge. The abundance of phylum Proteobacteria in the M-AnMBR was higher than that in the A-AnMBR, which should account for the higher removal of nutrients in the M-AnMBR. The faster membrane fouling would occur in the A-AnMBR due to the relatively high abundance of Bacteroidetes in the bulk sludge and cake sludge. Moreover, specific comparison down to the genus level showed that the dominant abundant bacterial genera were Candidate division OP8 norank and Anaerolineaceae uncultured in the cake sludge for M-AnMBR, and were VadinHA17 norank, WCHB1-69 norank, VadinBC27 wastewater-sludge group, and Synergistaceae uncultured in the cake sludge for A-AnMBR The different representative genera with the variation of the HRTs for the two bioreactors might indicate the different performance between the two AnMBRs.
1. Introduction
Anaerobic digestion can reduce the environmental threat of wastes and simultaneously produce energy in the form of biogas; therefore, it is an effective technology for waste treatment [1]. Membrane bioreactors (MBRs) have been increasingly and widely used for wastewater treatments in the last decade [2,3]. Due to the combination of anaerobic processes and membrane technologies, considerable attention has been devoted to the anaerobic membrane bioreactor (AnMBR) process for wastewater treatment [4]. AnMBRs have the ability to produce effluents similar in quality to those generated during aerobic treatment [5]. There are many cases in the literature investigating the efficiency of the AnMBR technology for wastewater treatment [5,6]. Lin et al. demonstrated the technical feasibility as well as economic feasibility of AnMBR treating wastewater [7].
It was believed that the performance of an anaerobic system has been reported to be tied closely with the microbial community structure [8]. Meanwhile, the bacterial community structure in MBR was significantly affected by the hydraulic retention time (HRT) [9]. It was obvious that anaerobic processes are strongly influenced by operational temperature; however, the heating of reactors demands energy and investment. Hence, anaerobic treatment has a great advantage at ambient temperatures during the process of wastewater treatment [10].
Membrane fouling is regarded as a major operational drawback for the membrane bioreactor (MBR) process, which has been researched thoroughly for aerobic MBR. The membrane fouling reduces permeate flux, which may require frequent membrane cleaning resulting in the decrease of the membrane lifespan. The microbial metabolites (extracellular polymeric substance (EPS) and soluble microbial product (SMP)) are identified to play significant roles in the membrane fouling formation [11,12].
The microbial community has a close relationship with the microbial metabolites [13,14], which had an effect on the membrane fouling in MBR. Meanwhile, the formation of membrane fouling in MBR is an intrinsically biological succession process of microbial communities on the membrane surface [15]. Therefore, the investigation on the microbial community is important to understand the membrane fouling phenomenon in MBR [13].
It had been revealed that some microorganisms in the cake sludge should be the result of the bulk sludge depositing [16]. However, the microbial communities in cake sludge and bulk sludge had a different correlation with membrane fouling during the MBR operation process [17,18]. It is interesting to identify the microbial communities in cake sludge and bulk sludge in AnMBR [19]. Nevertheless, the research of microbial community in bulk sludge and cake sludge is still limited in the AnMBR for wastewater treatment.
In the study, two AnMBRs with ring membrane modules treating municipal wastewater were operated on a laboratory scale under mesophilic and atmospheric conditions at different HRTs. The main objective was to evaluate long-term bacteria community by MiSeq high-throughput sequencing under mesophilic and atmospheric conditions with different HRTs. MiSeq high-throughput sequencing was carried out to investigate the microbial community structure in different environmental samples [20,21]. The community richness and diversity and species abundance were discussed emphatically to explore the relationship between MBR performance and the microbial community characters in bulk sludge and cake sludge. It is anticipated that this study could provide a new insight to understand the AnMBR treatment performance through researching the microbial community characters of bulk sludge and cake sludge under different HRTs.
2. Materials and Methods
2.1. AnMBRs Operation with Ring Membrane Modules
The two same AnMBRs were employed in this study and were operated under mesophilic temperature (35 °C) (M-AnMBR) and atmospheric temperature (25 °C) (A-AnMBR) (see Figure 1). The setup of the AnMBR had been introduced in a previous study [22]. The AnMBR was consisted of an anaerobic reactor with a working volume of 8 L and a ring membrane module. The AnMBR with a mixer (complete mixing) was fed with synthetic wastewater, and the effluent was collected with a suction mode of 4 min “on” and 1 min “off”. The transmembrane pressure (TMP) was monitored by a manometer, and once the TMP reached 30 kPa in any of the AnMBRs, the membrane modules in the two bioreactors were taken out and cleaned. The modules were reloaded into the bioreactors to run the next HRT after chemical cleaning. The AnMBRs were operated under three different HRTs conditions: 24 h, 12 h, and 6 h.
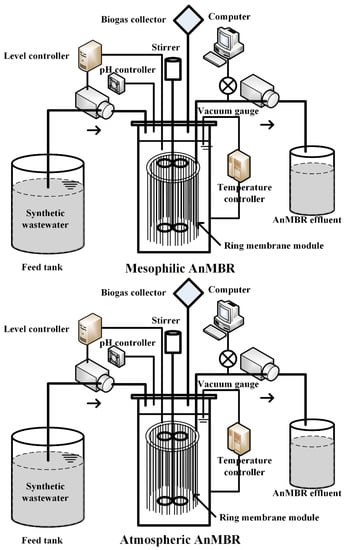
Figure 1.
Schematic of the mesophilic and atmospheric anaerobic membrane bioreactor and experimental setup.
2.2. Sludge Sample Collection
When the TMP increased to 30 kPa, the cake sludge formed on the membrane surface was carefully scraped off by a plastic sheet and simultaneously flushed with deionized (DI) water. After that, the membranes were soaked in 1% NaClO solution for chemical cleaning. The collected cake sludge samples were collected after being mixed on a magnetic blender. The bulk sludge samples were collected at the operation process under different HRTs. All samples were stored in a refrigerator (−20 °C) before further analysis.
2.3. Extraction of SMP and EPS
The supernatant was obtained by centrifuging biomass samples at 5000 rpm for 5 min, and then the SMP was collected after the supernatant was filtered through a 0.45 μm cellulose acetate membrane (Taoyuan, Haining, China). The loosely bound EPS (LB-EPS) and tightly bound EPS (TB-EPS) were extracted by two-step heating methods [23].
2.4. Microbial Community Analysis
According to a previous study [24], the bulk sludge and cake sludge obtained from the M-AnMBR and A-AnMBR were first processed for DNA extraction. The 16S rRNA genes were amplified from the extracted DNA with primer sets, 515F (5’-GTGCCAGCMGCCGCGG-3’) and 907R (5’-CCGTCAATTCMTTTRAGTTT-3’). Polymerase chain reaction (PCR) was performed in a thermocycler (ABI GeneAmp® 9700, Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: initial denaturing step of 3 min at 95 °C; followed by 25 cycles of denaturing (95 °C for 30 s), annealing (55 °C for 30 s), and elongation (72 °C for 45 s); and a final extension step at 72 °C for 10 min.
Purified PCR products were applied for sequencing on the Illumina Miseq platform (Illumina, San Diego, CA, USA). Rarefaction curves, Community richness Estimators of Chao1 and ACE, Community diversity Estimators of Shannon and Simpson, and the Good’s coverage were generated with MOTHUR for each sample. Representative sequences from each operational taxonomic unit (OTU) were assigned down to the phylum and class level, and the relative abundance of a given phylogenetic group was calculated. A hierarchical cluster (Heatmap) analysis and principal component analysis (PCA) were performed. Venn diagrams were obtained, and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) tree was constructed. The Miseq reads were also separately deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under accession numbers SRR8113740-SRR8113752.
2.5. Other Item Analysis
Chemical Oxygen Demands (COD) were determined according to the Chinese National Environmental Policy Act (NEPA) standard methods [25]. Protein and carbohydrate concentrations in the EPS and SMP samples were measured by the Lowry method and the phenol-sulphuric method, respectively. All the above analyses were performed in duplicate, and their average values were listed.
3. Results and Discussion
3.1. Performance of the AnMBRs
The treatment performance of the AnMBRs is shown in Table 1. The COD removal performed stable during the 189 days’ operation for M-AnMBR with the removal efficiencies of 91.09%, 93.51%, and 92.39%, respectively for HRT 24 h, 12 h, and 6 h. Compared to that of the M-AnMBR, the removal efficiency of the A-AnMBR was found to be lower under each HRT. It is interesting to observe that the highest removal efficiency both in the two bioreactors was occurred under HRT 12 h.

Table 1.
Wastewater treatment efficiencies of the anaerobic membrane bioreactors under mesophilic temperature (M-AnMBR) and atmospheric temperature (A-AnMBR) with different hydraulic retention times (HRTs).
It was seen from Figure 2 that the TMP always maintained lower level in the M-AnMBR and the faster membrane fouling occurred in the A-AnMBR under each HRT. In the A-AnMBR, the time for TMP reaching more than 0.030 MPa was 107 days, 52 days, and 30 days for HRT 24 h, 12 h, and 6 h, respectively. At HRT 24 h, the A-AnMBR was operated with a longer membrane fouling period compared to the other HRTs.
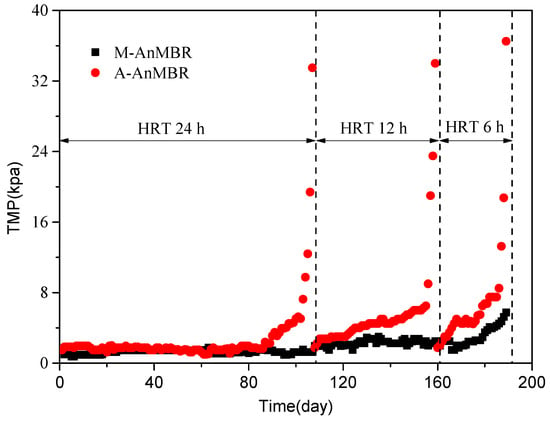
Figure 2.
The transmembrane pressure (TMP) profiles plotted as a function of time in the M-AnMBR and A-AnMBR.
It is widely accepted that membrane fouling in the membrane bioreactor is significantly affected by SMP and EPS [26]. Sludge cells in the floc matrix have a double layered EPS structure: LB-EPS diffused from TB-EPS surrounding the cells [27,28]. Carbohydrates and proteins as the major component of SMP, LB-EPS, and TB-EPS were analyzed, as depicted in Figure 3. It was noted that the concentrations of carbohydrates and proteins of LB-EPS and TB-EPS were increased obviously with the decrease of HRT in the two bioreactors, and the values of the M-AnMBR were much lower than that of the A-AnMBR. The different contents of SMP and EPS with the variation of the HRT in the two AnMBRs could be attributed to the variation of the microbial community structure, which plays an important role in the component of microbial metabolites.
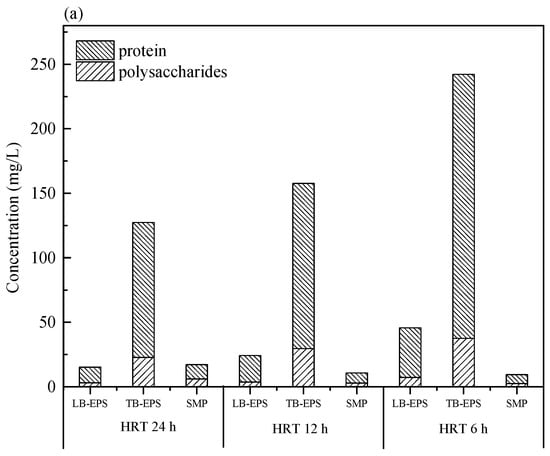
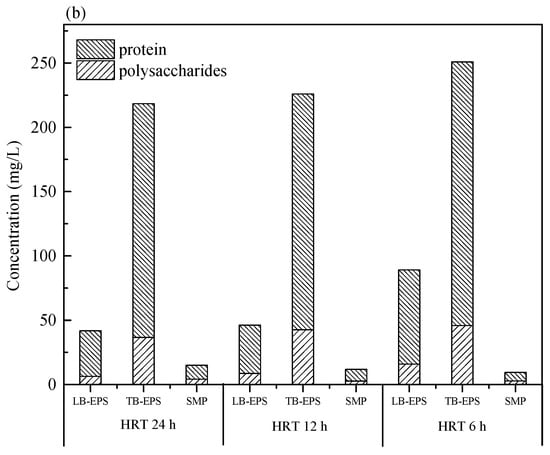
Figure 3.
Quantitative analysis of soluble microbial product (SMP) and extracellular polymeric substance (EPS) concentrations in the M-AnMBR (a) and A-AnMBR (b).
3.2. Microbial Richness and Diversity Evolution with the Variation of HRT in M-AnMBR and A-AnMBR
The MiSeq high-throughput sequencing was applied to analyze the microbial community structure in the bulk sludge and cake sludge samples of the AnMBRs under HRT 24 h, 12 h, and 6 h. The biomass samples were collected when the TMP reached 30 kPa, including bulk sludge samples from M-AnMBR at HRT 24 h (D1), HRT 12 h (D5), and HRT 6 h (D9) and from A-AnMBR at HRT 24 h (D2), HRT 12 h (D6), and HRT 6 h (D10) and cake sludge samples from M-AnMBR at HRT 24 h (D3), HRT 12 h (D7), and HRT 6 h (D11) and from A-AnMBR at HRT 24 h (D4), HRT 12 h (D8), and HRT 6 h (D12). There was also a single sample of the inoculative sludge named D13. Table 2 summarized the diversity estimators of Chao1, ACE and Shannon index, Simpson index, and Good’s coverage of sequencing for each sample.

Table 2.
Richness and diversity estimators of the bacterial communities of bulk sludge and cake sludge in the M-AnMBR and A-AnMBR.
In the M-AnMBR, the community richness of the bulk sludge with the decrease of the HRT kept a relatively constant level, and the community diversity of bulk sludge increased with the decrease of the HRT. For the cake sludge in the M-AnMBR, the community richness was the highest at HRT 24 h, and the community richness at HRT 12 h and 6 h were similar. The community diversity in the cake sludge in the M-AnMBR was the highest at HRT 24 h, and the community diversity at HRT 12 h was lower than that at HRT 6 h. For the M-AnMBR, it was somewhat reasonable that the increased loading has no significant effect on diversity and richness in the bulk sludge due to the lower organic loading rate of municipal wastewater, but the community diversity of bulk sludge increased with the increased loading. Meanwhile, the loading change had no obvious promoting effect upon the community richness and diversity of the cake sludge. On the contrary, the communities of the cake sludge indicated a slight drop in richness and diversity.
In the A-AnMBR, the richness and diversity of the bulk sludge were the highest for the HRT 12 h among the three HRTs; meanwhile, the richness and diversity of the bulk sludge at HRT 6 were higher than those at HRT 24. In the A-AnMBR, the richness and diversity of the cake sludge at HRT 24 h were similar to the community at HRT 12 h; however, the richness and diversity of the cake sludge at HRT 6 h were higher than those at HRT 24 h and 12 h. For the A-AnMBR, the highest richness and diversity of the bulk sludge for the HRT 12 h might be responsible for the highest removal efficiencies of the organic matter among the three HRTs. The increased loading has some significant effect on the community diversity and richness in the bulk sludge, and the community diversity of the bulk sludge and richness was first increased and then decreased along with the increasing of loading. Meanwhile, the loading change had an obvious promoting effect upon the community richness and diversity of the cake sludge, and the communities of the cake sludge indicated a slight increase in richness and diversity. The highest richness and diversity of the cake sludge for the HRT 6 h might be responsible for the highest membrane among the three HRTs.
In the M-AnMBR and A-AnMBR, the microbial community richness of the bulk sludge at each HRT were all lower than those of the cake sludge. The microbial community diversity of the bulk sludge at HRT 24 h and 6 h were lower than those of the cake sludge. However, at HRT 12 h, the diversity of the bulk sludge and cake sludge were similar in the A-AnMBR and the diversity of the bulk sludge was higher than that of cake sludge in the M-AnMBR. It is noteworthy that for the HRT 24 h, the richness of the cake sludge was similar in the A-AnMBR and M-AnMBR and the richness of bulk sludge was higher in the A-AnMBR compared to the M-AnMBR. However, the diversity of the cake sludge and bulk sludge was higher in the M-AnMBR compared to that in the A-AnMBR at HRT 24 h. The bulk sludge and cake sludge of A-AnMBR had the higher richness than those of M-AnMBR under HRT 12 h and HRT 6 h. The bulk sludge of A-AnMBR had a higher diversity than that of M-AnMBR under HRT 12 h, and the bulk sludge of A-AnMBR had a lower diversity than that of M-AnMBR at HRT 6 h. The cake sludge of A-AnMBR had the higher diversity than those of M-AnMBR under HRT 12 h and HRT 6 h.
The rarefaction curves were generated for each individual sludge sample, as shown in Figure S1. In the M-AnMBR, the microbial community richness of the bulk sludge and cake sludge at HRT 12 h and 6 h were lower than those in the A-AnMBR. At HRT 24 h, the richness of the bulk sludge and cake sludge in the M-AnMBR were similar with those in the A-AnMBR. This observation was in accordance with the results obtained by Chao1, ACE, Shannon, Simpson, and Coverage estimators (Table 2), indicating that the variation trend of the microbial community diversity and richness of the M-AnMBR was different with that of the A-AnMBR with different HRTs.
For the M-AnMBR and A-AnMBR, the loading change had a different effect on the community richness and diversity of the cake and bulk sludges. For the A-AnMBR, the increase in the organic loading rate was negative on COD removal, and therefore, COD removal efficiency decreased. For the M-AnMBR, the increase in the organic loading rate had little significant on the COD removal efficiency due to the higher microbial activity. This could be attributed to the different microbial activity between the M-AnMBR and A-AnMBR, and there was a lower microbial activity at 25 °C compared with 35 °C. With the change in HRTs, the distinct differences in the microbial richness and diversity between the two AnMBRs also indicated that the removal efficiencies of organic matter and membrane fouling in the two AnMBRs were different. The change in HRTs and different operation temperatures likely induced an environmental stress on the microbial communities, which subsequently influenced the community structure and composition.
3.3. Microbial Phylum and Class Identification with Different HRTs in M-AnMBR and A-AnMBR
Qualified reads were assigned to known phyla and class levels to characterize the bacterial communities (Figure 4). As seen from Figure 4a, all communities showed extremely high diversity, and 15 phyla were detected in all the sludge samples.
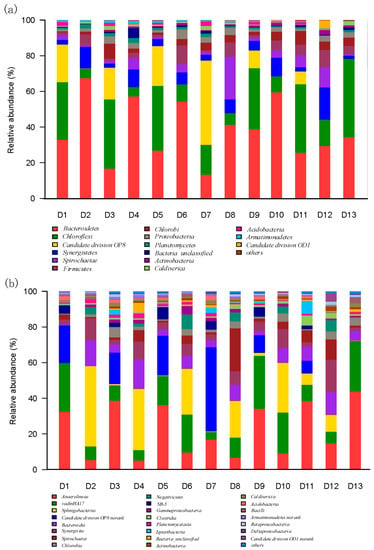
Figure 4.
The taxonomic compositions of bacterial communities at phyla (a) and class (b) levels in each sample retrieved from the Illumina-MiSeq sequencing.
In the M-AnMBR, Bacteroidetes and Chloroflexi were the main phyla for the microbial communities of bulk sludge under each HRT, and the sum of the two phyla were 65.26%, 63.14%, and 73.06% for HRT 24 h, 12 h, and 6 h, respectively. The abundance of Candidate division OP8 of bulk sludge in M-AnMBR at the HRT 24 h (21.00%) was similar with that at HRT 12 h (22.31%), whereas the abundance of Candidate division OP8 at HRT 6 h significantly reduced to 9.90%. In M-AnMBR, the abundances of Synergistetes, Chlorobi, Proteobacteria, and Actinobacteria of bulk sludge were increased with the reduction of HRT. The main microbial phyla of the cake sludge in M-AnMBR under different HRTs were Bacteroidetes, Chloroflexi, and Candidate division OP8, and the sum of the three phyla were 73.29%, 77.37%, and 71.20% for HRT 24 h, 12 h, and 6 h, respectively.
For the A-AnMBR, Bacteroidetes, Chloroflexi, Synergistetes, and Firmicutes were the main phyla in the bulk sludge under different HRTs, and the sum of the four main phyla were 91.23%, 81.23%, and 84.30% for HRT 24 h, 12 h, and 6 h, respectively. The proportion of Bacteroidetes was the largest in the bulk sludge of the A-AnMBR, and it was 67.38%, 54.41%, and 59.38% for HRT 24 h, 12 h, and 6 h, respectively. There were five major phyla in the cake sludge of the A-AnMBR, and the dominant phyla were Bacteroidetes, Chloroflexi, Synergistetes, Firmicutes, and Spirochaetae. The proportion of the five phyla in the cake sludge were 81.72%, 87.55%, and 83.28% for HRT 24 h, 12 h, and 6 h, respectively.
It was found that most of the sequence reads were affiliated to six major phyla: Bacteroidetes, Chloroflexi, Synergistetes, Spirochaetae, Firmicutes, and Chlorobi in all samples. The majority of these phyla were ubiquitous in anaerobic bioreactors [19,29]. Meanwhile, it is interesting to find that the phylum of Candidate division OP8 was only major in the M-AnMBR, and its abundance was reached up to 47.25% in the cake sludge of the M-AnMBR at HRT 12 h. However, there was little abundance of Candidate division OP8 in the A-AnMBR. The abundance of the main phyla in the two bioreactors also have obvious difference. Contrasting the biological community structures of the two reactors, the abundance of phylum Proteobacteria in the M-AnMBR was higher than that in the A-AnMBR. Proteobacteria includes a variety of functionally important bacteria for organic pollutants removal [30,31], which should account for the higher removal of nutrients in the M-AnMBR. The abundances of Bacteroidetes and Synergistetes in the M-AnMBR were significantly lower than that of the A-AnMBR under each HRT both in the bulk sludge and cake sludge. It was reported that Bacteroidetes could potentially release more proteinaceous EPS, which was beneficial to its colonization [32,33]. Meanwhile, Bacteroidetes bacterium possesses fimbriae, which could help bacterium attach to the surfaces and may contribute to Bacteroidetes stable proliferation on the supporting material [34]. Hence, Bacteroidetes were highly enriched on the membrane surface and enhanced the bacteria adhere to the membrane. Therefore, the faster membrane fouling would occur in the A-AnMBR due to the relatively high abundance of Bacteroidetes in the bulk sludge and cake sludge.
Further comparison of the phyla down to the class level was conducted to reveal more information on the microbial community evolution between M-AnMBR and A-AnMBR. The bacterial communities based on the class-level identification is illustrated in Figure 4b. A total of 23 bacterial classes were found in all thirteen sludge samples. It was observed that the main community structure was obviously different in the two bioreactors. Anaerolineae, vadinHA17, and Candidate division OP8 norank belonging to Chloroflexi, Bacteroidetes, and Candidate division OP8, respectively, were the three major classes and accounted for 80.85%, 74.81%, and 73.75% of the total population in the bulk sludge of the M-AnMBR for 24 h, 12 h, and 6 h, respectively. The class Anaerolineae were anaerobic bacteria that was involved in carbohydrate decomposition in anaerobic systems [35]. Candidate division OP8 was reported as important members of thermophilic microbial communities related to the anaerobic degradation of hydrocarbons [36,37,38].
There were four main classes in the bulk sludge of the A-AnMBR: vadinHA17, Sphingobacteriia, and Bacteroidia belong to the Bacteroidetes, and Synergistia belongs to Synergistetes. Sphingobacteriia was the largest part, and it accounted for 45.02%, 25.6%, and 27.3% for 24 h, 12 h, and 6 h, respectively. Typically, most bacteria that can be attributed to Bacteroidetes can produce various lytic enzymes and acetic acid during the organic materials decomposition [39,40], and thus the contribution of the classes in this study was probably associated with organic matter removal. Synergistia (phylum Synergistetes) was very abundant, and the class Synergistia includes numerous bacteria that can efficiently decompose complex organic matters [41,42,43].
In the cake sludge of the M-AnMBR, the main classes were similar with those in the bulk sludge, but the proportion of these classes was different, and the vadinHA17 in the cake sludge was apparently lower than that in the bulk sludge. For the cake sludge of the A-AnMBR, the classes Spirochaetes became the dominant besides the four main classes in the bulk sludge. Meanwhile, the proportion of Sphingobacteriia reduced a lot, and it was 34.28%, 20.64%, and 9.4% for HRT 24 h, 12 h, and 6 h, respectively, which was reduced with the HRT reduction. The abundance of Bacteroidia and Spirochaetes in the cake sludge of the A-AnMBR was respectively higher than that in the bulk sludge. Especially, Spirochaetes were increased from 0.68%, 4.62%, and 4.15% in the bulk sludge to 6.5%, 24.4%, and 11.39% in the cake sludge for HRT 24 h, 12 h, and 6 h, respectively. It has been known that Spirochaetes are chemoheterotrophic and have a distinctive spiral shape [44]. The Spirochaetes were found in the methanogenic anaerobic reactor [45], and they were anticipated to retrieve energy through the fermentation of either carbohydrates or amino acids [46].
For the bulk sludge and cake sludge from the M-AnMBR, the bacteria belonging to the class Anaerolineae and Candidate division OP8 norank were higher than those for the bulk sludge and cake sludge from the A-AnMBR under each HRT, but Sphingobacteriia, Bacteroidia, and Synergistia were lower than those for the bulk sludge and cake sludge from the A-AnMBR under each HRT. The abundances of Spirochaetes in the bulk sludge from the M-AnMBR was higher than that in the bulk sludge from the A-AnMBR at HRT 24 h, but for HRT 12 h and 6 h, the abundances of Spirochaetes in the bulk sludge from the M-AnMBR were lower. However, the abundance of Spirochaetes always was lower for the cake sludge from the M-AnMBR than that for the cake sludge from the A-AnMBR under the corresponding HRT. Thus, the different treating performances would occur between the M-AnMBR and A-AnMBR due to the different microbial communities in the bulk sludge and cake sludge with different HRTs. In order to identify the different compositions of these microbial communities, an investigation down to the genus level was needed to further elucidate the functions of the microbial communities involved in the AnMBR process.
3.4. Microbial Genera Evolution with the Variation of HRT in M-AnMBR and A-AnMBR
The heatmap shows the relative abundance of different bacteria genera found in the samples, as presented in Figure 5. It can be seen that the representative genera were different between the two bioreactors.
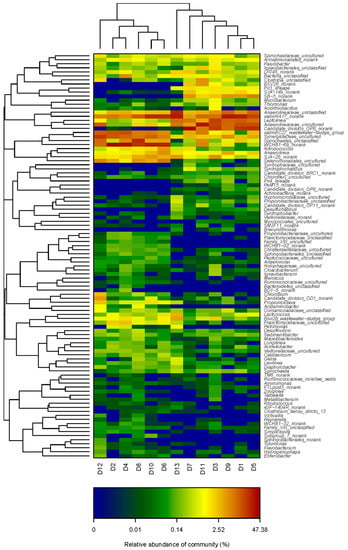
Figure 5.
Heatmaps showed the percentages of the abundant species at the genus level within the bacterial communities of the bulk sludge and cake sludge in the M-AnMBR and A-AnMBR.
For the bulk sludge from the M-AnMBR, the bacteria belonging to the genera Anaerolineaceae unclassified, VadinHA17 norank, Leptolinea, Anaerolineaceae uncultured, and Candidate division OP8 norank were dominant in all the HRTs. For the cake sludge from the M-AnMBR, Leptolinea, Anaerolineaceae uncultured, and Candidate division OP8 norank were dominant at HRT 24 h, and the relative abundances of them were higher than those in the bulk sludge. Candidate division OP8 norank and Anaerolineaceae uncultured became the most dominant bacterial community for HRT 12 h and 6 h, respectively, for the cake sludge from the M-AnMBR. Meanwhile, in the M-AnMBR, the relative abundances of Candidate division OP8 norank and Anaerolineaceae uncultured in the cake sludge were higher than those in the bulk sludge.
In the bulk sludge from the A-AnMBR, the microbial community was also different with the variation of the HRTs. WCHB1-69 norank, VadinHA17 norank, VadinBC27 wastewater-sludge group, and Synergistaceae uncultured were predominant in all HRTs. Spirochaetes unclassified became the dominant with the decrease of the HRT. For the cake sludge from the A-AnMBR, VadinHA17 norank, WCHB1-69 norank, VadinBC27 wastewater-sludge group, and Synergistaceae uncultured were dominant; however, the relative abundances of VadinHA17 norank and WCHB1-69 norank in the cake sludge were lower than that in the bulk sludge. The relative abundances of Spirochaetes unclassified and Synergistaceae uncultured in the cake sludge from the A-AnMBR were higher than that in the bulk sludge. Anaerolineaceae uncultured became the dominant bacterial community with the decrease of the HRT in the cake sludge from the A-AnMBR.
Obviously, it could be found that the dominant abundant bacterial genera were Candidate division OP8 norank and Anaerolineaceae uncultured in the cake sludge for the M-AnMBR, and the dominant abundant bacterial genera were VadinHA17 norank, WCHB1-69 norank, VadinBC27 wastewater-sludge group, and Synergistaceae uncultured in the cake sludge for the A-AnMBR. The results indicated that the microbial communities were quite different for the cake sludge between the two AnMBRs. The different dominant abundant bacterial genus in the cake sludge might be responsible for the different membrane fouling in the M-AnMBR and A-AnMBR.
The bacterial communities were clearly divided into two groups based on PCA and UPGMA from the Unifrac and Mothur analyses (Figure S2). PCA indicated that principal components 1 and 2 explained 52.06% and 20.82% of the total community variations, respectively. The results indicate there was a divergence between the two bioreactors, suggesting that these samples exhibited different structures. The bulk sludge samples in all the HRTs and the cake sludge samples at HRT 24 h and HRT 6 h were clustered together in the M-AnMBR, which showed that the bacterial community compositions were similar at M-AnMBR, whereas the cake sludge samples at HRT 12 h was well separated from the other sludge samples. In the A-AnMBR, the bacterial community compositions of the bulk sludge and cake sludge were similar.
The UPGMA tree revealed two different clusters: group 1 comprised all the samples (bulk sludge and cake sludge) from the M-AnMBR and group 2 comprised all the samples from the A-AnMBR. This indicates that the bacterial communities differed significantly between the two bioreactors; however, the bacterial communities were similar within the same bioreactors.
Venn diagrams were obtained to show the shared and unique OTUs to make further community-wide comparisons (Figure S3). Figure S3a1 showed the shared and unique OTUs of the bulk sludge in the M-AnMBR under the three HRTs and the inoculation sludge (D13). The number of the total observed OTUs in the M-AnMBR under the three HRTs and the inoculation sludge was 291, and 64 OTUs (about 22.0% of the total) were shared in the M-AnMBR under the three HRTs and the inoculation sludge. The number of total observed OTUs in the A-AnMBR under the three HRTs and the inoculation sludge was 314, and there were 70 OTUs (about 22.3% of the total) shared in the A-AnMBR under the three HRTs and the inoculation sludge (Figure S3a2). The shared and unique OTUs of the cake sludge in the M-AnMBR and A-AnMBR under the three HRTs were showed in Figure S3b1,b2, and 122 OTUs (about 40.5% of the total) and 163 OTUs (about 52.1% of the total) were shared under the three HRTs in the M-AnMBR and A-AnMBR, respectively.
The Venn diagrams, PCA, and UPGMA indicated that the HRT affected the bacterial community abundance in the M-AnMBR and A-AnMBR. The changes of the dominant bacterial community in the M-AnMBR and A-AnMBR could explain the variances in performance between the M-AnMBR and A-AnMBR. The results from the Illumina-MiSeq sequencing further advanced our knowledge on the microbial compositions of the cake sludge and bulk sludge in the M-AnMBR and A-AnMBR, which may provide a better understanding of the AnMBR operation mechanisms and the improvement of the AnMBR performance.
4. Conclusions
Mesophilic and atmospheric anaerobic membrane bioreactors (AnMBR) were operated for treating municipal wastewater with different HRTs. Compared to the M-AnMBR, the removal efficiency of the A-AnMBR was found to be lower, and the faster membrane fouling occurred in A-AnMBR under corresponding hydraulic retention times (HRT). For the M-AnMBR and A-AnMBR, the HRT change had different effects on the community richness and diversity of the cake and bulk sludge. This could be attributed to the different microbial activity between the M-AnMBR and A-AnMBR. The Anaerolineae, vadinHA17, and Candidate division OP8 norank were the major classes in the bulk sludge of the M-AnMBR, and the main classes in the bulk sludge of the A-AnMBR were vadinHA17, Sphingobacteriia, Bacteroidia, and Synergistia. Moreover, at the genus level, the dominant abundant bacterial genera were Candidate division OP8 norank and Anaerolineaceae uncultured in the cake sludge for the M-AnMBR, and the dominant abundant bacterial genera were VadinHA17 norank, WCHB1-69 norank, VadinBC27 wastewater-sludge group, and Synergistaceae uncultured in the cake sludge for the A-AnMBR. The distinct differences in the microbial community structures of the bulk sludge might indicate that the removal efficiencies of organic matter in the two AnMBRs were different with different HRTs. The different dominant abundant bacterial genus in the cake sludge might be responsible for the different membrane fouling in the M-AnMBR and A-AnMBR.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4441/11/1/160/s1, Figure S1: The rarefaction curves base on the Illumina-MiSeq sequencing of the bulk sludge and cake sludge bacterial communities in the M-AnMBR and A-AnMBR. The operational taxonomic units (OTUs) were defined by 3% distances; Figure S2: The principal component analysis (PCA) of the bacterial communities from the bulk sludge and cake sludge in the M-AnMBR and A-AnMBR based on the Illumina-MiSeq sequencing of the 16S rRNA gene (a) and the unweighted pair group method with arithmetic mean (UPGMA) tree showing relationships between samples according to bacterial profile in the M-AnMBR and A-AnMBR (b). The operational taxonomic units (OTUs) were determined based on 97% similarity of reads, Figure S3: Venn diagrams showing the overlap of OTUs from the bulk sludge (a) and cake sludge (b) in the M-AnMBR (1) and A-AnMBR (2).
Author Contributions
Data curation, Z.G.; formal analysis, C.J.; investigation, Y.D. and Z.L. (Zhenlin Liang); writing—original draft, Z.L. (Zhipeng Li); writing—review and editing, X.H.
Funding
This study was funded by the National Natural Science Fund of China (No. 51408158), the China Postdoctoral Science Foundation (No. 2017M612278), Shandong Provincial Natural Science Foundation, China (No. ZR2017PEE008 and No. ZR2017MEE020), the Fundamental Research Funds for the Central Universities (No. HIT.NSRIF.2016098) and the scientific research foundation of Harbin institute of technology at Weihai (HIT(WH)201403).
Acknowledgments
The authors are grateful to the editors and two anonymous reviewers for their constructive comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Luo, G.; Wang, W.; Angelidaki, I. Anaerobic Digestion for Simultaneous Sewage Sludge Treatment and CO Biomethanation: Process Performance and Microbial Ecology. Environ. Sci. Technol. 2013, 47, 10685–10693. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.-K.; Kim, J.-H.; Ahn, C.H.; Lee, C.-H.; Song, J.-Y.; Ra, Y.-H. Effect of disintegrated sludge recycling on membrane permeability in a membrane bioreactor combined with a turbulent jet flow ozone contactor. Water Res. 2010, 44, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Chae, S.-R.; Drews, A.; Kraume, M.; Shin, H.-S.; Yang, F. Recent advances in membrane bioreactors (MBRs): Membrane fouling and membrane material. Water Res. 2009, 43, 1489–1512. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.J.; Qu, X.; Leung, K.T.; Liao, B.Q. Influence of temperature and temperature shock on sludge properties, cake layer structure, and membrane fouling in a submerged anaerobic membrane bioreactor. J. Membr. Sci. 2012, 421–422, 131–144. [Google Scholar] [CrossRef]
- Smith, A.L.; Stadler, L.B.; Love, N.G.; Skerlos, S.J.; Raskin, L. Perspectives on anaerobic membrane bioreactor treatment of domestic wastewater: A critical review. Bioresour. Technol. 2012, 122, 149–159. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Q.; Zheng, S.S.; Wu, D.L. Anaerobic membrane bioreactor (AnMBR) for bamboo industry wastewater treatment. Bioresour. Technol. 2013, 149, 292–300. [Google Scholar] [CrossRef]
- Lin, H.; Chen, J.; Wang, F.; Ding, L.; Hong, H. Feasibility evaluation of submerged anaerobic membrane bioreactor for municipal secondary wastewater treatment. Desalination 2011, 280, 120–126. [Google Scholar] [CrossRef]
- Mcmahon, K.D.; Stroot, P.G.; Mackie, R.I.; Raskin, L. Anaerobic codigestion of municipal solid waste and biosolids under various mixing conditions—II: Microbial population dynamics. Water Res. 2001, 35, 1804–1816. [Google Scholar] [CrossRef]
- Gao, D.W.; Tao, Y.; An, R. Digested sewage treatment using membrane-based process at different hydraulic retention times. Desalination 2012, 286, 187–192. [Google Scholar] [CrossRef]
- Martinez-Sosa, D.; Helmreich, B.; Netter, T.; Paris, S.; Bischof, F.; Horn, H. Anaerobic submerged membrane bioreactor (AnSMBR) for municipal wastewater treatment under mesophilic and psychrophilic temperature conditions. Bioresour. Technol. 2011, 102, 10377–10385. [Google Scholar] [CrossRef]
- Gao, W.J.J.; Lin, H.J.; Leung, K.T.; Liao, B.Q. Influence of elevated pH shocks on the performance of a submerged anaerobic membrane bioreactor. Process Biochem. 2010, 45, 1279–1287. [Google Scholar] [CrossRef]
- Huang, X.; Liu, R.; Qian, Y. Behaviour of soluble microbial products in a membrane bioreactor. Process Biochem. 2000, 36, 401–406. [Google Scholar] [CrossRef]
- Gao, D.W.; Wen, Z.D.; Li, B.; Liang, H. Microbial community structure characteristics associated membrane fouling in A/O-MBR system. Bioresour. Technol. 2014, 154, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Yeon, K.M.; Cheong, W.S.; Oh, H.S.; Lee, W.N.; Hwang, B.K.; Lee, C.H.; Beyenal, H.; Lewandowski, Z. Quorum Sensing: A New Biofouling Control Paradigm in a Membrane Bioreactor for Advanced Wastewater Treatment. Environ. Sci. Technol. 2009, 43, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kim, S.; Yeon, K.M.; Sang, B.I.; Chun, J.; Lee, C.H. Correlation between microbial community structure and biofouling in a laboratory scale membrane bioreactor with synthetic wastewater. Desalination 2012, 287, 209–215. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Wu, Z.; Wei, T.; Lu, F.; Tong, J.; Mai, S. Membrane fouling in an anaerobic dynamic membrane bioreactor (AnDMBR) for municipal wastewater treatment: Characteristics of membrane foulants and bulk sludge. Process Biochem. 2011, 46, 1538–1544. [Google Scholar] [CrossRef]
- Miura, Y.; Watanbe, Y.; Okabe, S. Membrane biofouling in pilot-scale membrane bioreactors (MBRs) treating municipal wastewater: Impact of biofilm formation. Environ. Sci. Technol. 2007, 41, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.N.; De Wever, H.; Diels, L. Diverse and Distinct Bacterial Communities Induced Biofilm Fouling in Membrane Bioreactors Operated under Different Conditions. Environ. Sci. Technol. 2008, 42, 8360–8366. [Google Scholar] [CrossRef]
- Ma, J.X.; Wang, Z.W.; Zou, X.X.; Feng, J.J.; Wu, Z.C. Microbial communities in an anaerobic dynamic membrane bioreactor (AnDMBR) for municipal wastewater treatment: Comparison of bulk sludge and cake layer. Process Biochem. 2013, 48, 510–516. [Google Scholar] [CrossRef]
- Liu, L.; Li, T.; Wei, X.; Jiang, B.; Fang, P. Effects of a nutrient additive on the density of functional bacteria and the microbial community structure of bioorganic fertilizer. Bioresour. Technol. 2014, 172, 328–334. [Google Scholar] [CrossRef]
- Liang, S.; Gliniewicz, K.; Mendes-Soares, H.; Settles, M.L.; Forney, L.J.; Coats, E.R.; McDonald, A.G. Comparative analysis of microbial community of novel lactic acid fermentation inoculated with different undefined mixed cultures. Bioresour. Technol. 2015, 179, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tian, Y.; Li, Z.P.; Liu, F.; You, H. Characterization of organic membrane foulants in a forward osmosis membrane bioreactor treating anaerobic membrane bioreactor effluent. Bioresour. Technol. 2014, 167, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Yang, S.F. Influence of loosely bound extracellular polymeric substances (EPS) on the flocculation, sedimentation and dewaterability of activated sludge. Water Res. 2007, 41, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tian, Y.; Liu, J.; Li, N.; Zhang, J.; Zuo, W.; Li, Z. Investigation of microbial structure and composition involved in membrane fouling in the forward osmosis membrane bioreactor treating anaerobic bioreactor effluent. Chem. Eng. J. 2016, 286, 198–207. [Google Scholar] [CrossRef]
- CEPB (Chinese Environmental Science Press). Standard Methods for Examination of Water and Wastewater, 4th ed.; Chinese Environmental Science Press: Beijing, China, 2002. [Google Scholar]
- Wang, Z.W.; Wu, Z.C. A Review of Membrane Fouling in MBRs: Characteristics and Role of Sludge Cake Formed on Membrane Surfaces. Sep. Sci. Technol. 2009, 44, 3571–3596. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. Relevance of microbial extracellular polymeric substances (EPSs)—Part I: Structural and ecological aspects. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Sheng, G.P.; Luo, H.W.; Zhang, F.; Yuan, S.J.; Xu, J.; Zeng, R.J.; Wu, J.G.; Yu, H.Q. Contribution of Extracellular Polymeric Substances (EPS) to the Sludge Aggregation. Environ. Sci. Technol. 2010, 44, 4355–4360. [Google Scholar] [CrossRef]
- Xie, Z.F.; Wang, Z.W.; Wang, Q.Y.; Zhu, C.W.; Wu, Z.C. An anaerobic dynamic membrane bioreactor (AnDMBR) for landfill leachate treatment: Performance and microbial community identification. Bioresour. Technol. 2014, 161, 29–39. [Google Scholar] [CrossRef]
- Schmidt, I.; Sliekers, O.; Schmid, M.; Bock, E.; Fuerst, J.; Kuenen, J.G.; Jetten, M.S.M.; Strous, M. New concepts of microbial treatment processes for the nitrogen removal in wastewater. FEMS Microbiol. Rev. 2003, 27, 481–492. [Google Scholar] [CrossRef]
- Nogueira, R.; Melo, L.F.; Purkhold, U.; Wuertz, S.; Wagner, M. Nitrifying and heterotrophic population dynamics in biofilm reactors: Effects of hydraulic retention time and the presence of organic carbon. Water Res. 2002, 36, 469–481. [Google Scholar] [CrossRef]
- Gao, D.W.; Zhang, T.; Tang, C.Y.Y.; Wu, W.M.; Wong, C.Y.; Lee, Y.H.; Yeh, D.H.; Criddle, C.S. Membrane fouling in an anaerobic membrane bioreactor: Differences in relative abundance of bacterial species in the membrane foulant layer and in suspension. J. Membr. Sci. 2010, 364, 331–338. [Google Scholar] [CrossRef]
- Yu, Z.Y.; Wen, X.H.; Xu, M.L.; Huang, X. Characteristics of extracellular polymeric substances and bacterial communities in an anaerobic membrane bioreactor coupled with online ultrasound equipment. Bioresour. Technol. 2012, 117, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D.; Li, J.; Guo, P.; Li, P.P.; Suo, Y.L.; Wang, X.F.; Cui, Z.J. Dynamic transition of microbial communities in response to acidification in fixed-bed anaerobic baffled reactors (FABR) of two different flow directions. Bioresour. Technol. 2011, 102, 4703–4711. [Google Scholar] [CrossRef] [PubMed]
- Narihiro, T.; Terada, T.; Ohashi, A.; Kamagata, Y.; Nakamura, K.; Sekiguchi, Y. Quantitative detection of previously characterized syntrophic bacteria in anaerobic wastewater treatment systems by sequence-specific rRNA cleavage method. Water Res. 2012, 46, 2167–2175. [Google Scholar] [CrossRef]
- Chen, C.L.; Macarie, H.; Ramirez, I.; Olmos, A.; Ong, S.L.; Monroy, O.; Liu, W.T. Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 2004, 150, 3429–3440. [Google Scholar] [CrossRef]
- Fang, H.H.P.; Liang, D.W.; Zhang, T.; Liu, Y. Anaerobic treatment of phenol in wastewater under thermophilic condition. Water Res. 2006, 40, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Mbadinga, S.M.; Li, K.P.; Zhou, L.; Wang, L.Y.; Yang, S.Z.; Liu, J.F.; Gu, J.D.; Mu, B.Z. Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 2012, 96, 531–542. [Google Scholar] [CrossRef]
- Chen, S.Y.; Dong, X.Z. Proteiniphilum acetatigenes gen. nov., sp nov., from a UASB reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 2005, 55, 2257–2261. [Google Scholar] [CrossRef]
- Riviere, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef]
- Wirth, R.; Kovács, E.; Maróti, G.; Bagi, Z.; Rákhely, G.; Kovács, K.L. Characterization of a biogas-producing microbial community by short-read next generation DNA sequencing. Biotechnol. Biofuels 2012, 5, 1–16. [Google Scholar] [CrossRef]
- Ito, T.; Yoshiguchi, K.; Ariesyady, H.D.; Okabe, S. Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J. 2011, 5, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.M.; Kim, J.H.; Ha, J.H.; Park, J.M. Bacterial and methanogenic archaeal communities during the single-stage anaerobic digestion of high-strength food wastewater. Bioresour. Technol. 2014, 165, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Chon, D.H.; Kim, H.S.; Park, C. Investigation of bacterial community in activated sludge with an anaerobic side-stream reactor (ASSR) to decrease the generation of excess sludge. Water Res. 2012, 46, 4292–4300. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.G.; Han, G.; Lim, J.; Lee, C.; Hwang, S. A comprehensive microbial insight into two-stage anaerobic digestion of food waste-recycling wastewater. Water Res. 2010, 44, 4838–4849. [Google Scholar] [CrossRef] [PubMed]
- Delbes, C.; Moletta, R.; Godon, J.J. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction—Single-strand conformation polymorphism analysis. Environ. Microbiol. 2000, 2, 506–515. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).