Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites and Sample Collection
2.2. Chemical Analysis of Water and Sediment Samples
2.3. Determination of Potential Denitrification Rate and Potential Nitrification Rate of Sediment
2.4. DNA Extraction and qPCR
2.5. Statistical Analysis
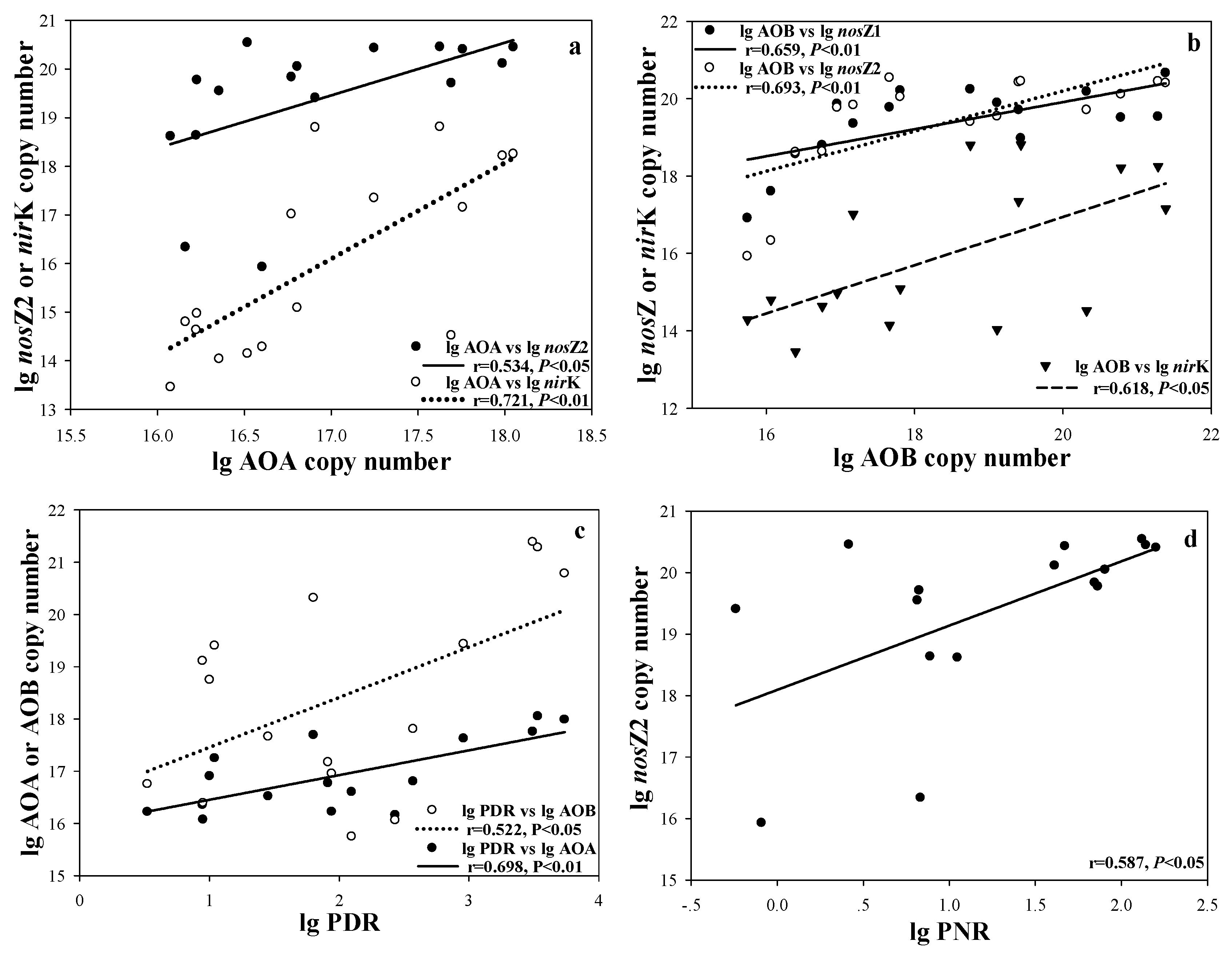
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
List of Abbreviations.
| CND | Coupled nitrification and denitrification |
| TN | Total Nitrogen |
| DTN | Dissolved Total Nitrogen |
| DON | Dissolved Organic Nitrogen |
| NH4+-N | Ammonium (nitrogen) |
| NO3−-N | Nitrate (nitrogen) |
| NO2−-N | Nitrite (nitrogen) |
| TP | Total Phosphorus |
| DTP | Dissolved Total Phosphorus |
| SRP | Soluble Reactive Phosphorus |
| Fe(OOH)~P | Iron-bound phosphorus |
| CaCO3~P | Calcium-bound phosphorus |
| ASOP | Acid-Soluble Organic Phosphorus |
| Palk | Hot NaOH-extractable organic phosphorus |
| EEA | Extracellular Enzyme Activity |
| APA | Alkaline Phosphatase Activity |
| GLU | β-D-glucosidase activity |
| LAP | Leucine aminopeptidase activity |
| Chl a | Chlorophyll a |
| TSI | Trophic State Index |
| PDR | Potential Denitrification Rate |
| PNR | Potential Nitrification Rate |
| AOA | Ammonia-Oxidizing Archaea |
| AOB | Ammonia-Oxidizing Bacteria |
| nir | Nitrite reductase |
| nos | Nitrous oxide reductase |
References
- Jie, L.; SiFan, Z.; Lin, X. Effect of water bloom on the nitrogen transformation and the relevant bacteria. Environ. Sci. 2016, 6, 2164–2170. [Google Scholar] [CrossRef]
- Abell, G.C.; Revill, A.T.; Smith, C.; Bissett, A.P.; Volkman, J.K.; Robert, S.S. Archaeal ammonia oxidizers and nirS-type denitrifiers dominate sediment nitrifying and denitrifying populations in a subtropical macrotidal estuary. ISME J. 2009, 4, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Li, J.; Wang, R.; Li, J.Y.; Zhang, Z.Q. Tracking composition and dynamics of nitrification and denitrification microbial community in a biofilm reactor by PCR-DGGE and combining FISH with flow cytometry. Biochem. Eng. J. 2010, 49, 370–378. [Google Scholar] [CrossRef]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. P. Natl. Acad. Sci. USA 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.H.; Zhong, J.C.; Zheng, X.L.; Fan, C.X.; Yu, J.H.; Zhong, W.H. N2O Fluxes and Rates of Nitrification and Denitrification at the Sediment–Water Interface in Taihu Lake, China. Water 2018, 10, 911. [Google Scholar] [CrossRef]
- Hill, A.R. Denitrification in the Nitrogen Budget of a River Ecosystem. Nature 1979, 281, 291–292. [Google Scholar] [CrossRef]
- Wrage, N.; Velthof, G.L.; Beusichem, M.L.V.; Oenema, O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 2001, 33, 1–1732. [Google Scholar] [CrossRef]
- Kremen, A.; Bear, J.; Shavit, U.; Shaviv, A. Model demonstrating the potential for coupled nitrification denitrification in soil aggregates. Environ. Sci. Technol. 2005, 39, 4180–4188. [Google Scholar] [CrossRef] [PubMed]
- Konneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Heylen, K.; Gevers, D.; Vanparys, B.; Wittebolle, L.; Geets, J.; Boon, N.; De Vos, P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 2010, 8, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Ginger, L.J.; Zimmer, K.D.; Herwig, B.R.; Hanson, M.A.; Hobbs, W.O.; Small, G.E.; Cotner, J.B. Watershed versus within-lake drivers of nitrogen: Phosphorus dynamics in shallow lakes. Ecol. Appl. 2017, 27, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Lisa, J.A.; Song, B.; Tobias, C.R.; Hines, D.E. Genetic and biogeochemical investigation of sedimentary nitrogen cycling communities responding to tidal and seasonal dynamics in Cape Fear River Estuary. Estuar. Coast. Shelf Sci. 2015, 167, A313–A323. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Bae, H.S.; Reddy, K.R.; Ogram, A. Distributions, abundances and activities of microbes associated with the nitrogen cycle in riparian and stream sediments of a river tributary. Water Res. 2016, 106, 51–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Zhao, Y.; Xie, S.; Li, J.Y. Implication of Nitrifying and Denitrifying Bacteria for Nitrogen Removal in a Shallow Lake. Clean Soil Air, Water 2017, 45, 1500319. [Google Scholar] [CrossRef]
- He, Q.L.; Chen, L.; Zhang, S.J.; Wang, L.; Liang, J.W.; Xia, W.H.; Wang, H.Y.; Zhou, J.P. Simultaneous nitrification, denitrification and phosphorus removal in aerobic granular sequencing batch reactors with high aeration intensity: Impact of aeration time. Biores Technol. 2018, 263, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.F.; Yu, H.J. Study on the technology of shortcut nitrification-denitrification simultaneous phosphorus removal. Advan. Mater. Res. 2013, 864–867, 1503–1508. [Google Scholar] [CrossRef]
- Yu, D.S.; Yuan, M.F.; Wang, X.X.; Chen, G.H.; Zhen, J.Y.; Du, S.M.; Zhang, F. Simultaneous Nitrogen and Phosphorus Removal Characteristics of An Anaerobic/Aerobic Operated SPNDPR System Treating Low C/N Urban Sewage. Environ. Sci. 2018. [Google Scholar] [CrossRef]
- Preisendorfer, R.W. Secchi Disk Science: Visual Optics of Natural Waters. Limnol. Oceanogr. 1986, 31, 909–926. [Google Scholar] [CrossRef]
- Jespersen, A.M.; Christoffersen, K. Measurements of chlorophyll-a from phytoplankton using ethanol as extraction solvent. Arch. Hydrobiol. 1987, 109, 445–454. [Google Scholar]
- Carlson, R.E. Trophic State Index for Lakes. Limnol. Oceanogr. Meth. 1977, 22, 361–369. [Google Scholar] [CrossRef]
- Boetius, A.; Lochte, K. Regulation of microbial enzymatic degradation of OM in deep-sea sediments. Mar. Ecol. Prog. 1994, 104, 299–307. [Google Scholar] [CrossRef]
- Golterman, H.L. Fractionation of sediment phosphate with chelating compounds: A simplification, and comparison with other methods. Hydrobiologia 1996, 335, 87–95. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, P. A modified single solution method of the determination of phosphate in natural waters. Anal. Chimica Acta 1962, 27, 1–36. [Google Scholar] [CrossRef]
- Jha, P.K.; Minagawa, M. Assessment of denitrification process in lower Ishikari river system, Japan. Chemosphere. 2013, 93, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.L.; Zhang, F.S.; Lu, Y.H. Linking plant identity and interspecific competition to soil nitrogen cycling through ammonia oxidizer communities. Soil Biol. Biochem. 2011, 43, 46–54. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Winder, R.S. Quantification of Nitrogen Reductase and Nitrite Reductase Genes in Soil of Thinned and Clear-Cut Douglas-Fir Stands by Using Real-Time PCR. Appl. Environ. Microb. 2010, 76, 7116–7125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallin, S.; Lindgren, P.E. PCR detection of genes encoding nitrile reductase in denitrifying bacteria. Appl. Environ. Microb. 1999, 65, 1652–1657. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microb. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Nicolaisen, M.H.; Ramsing, N.B. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Meth. 2002, 50, 189–203. [Google Scholar] [CrossRef]
- Li, B.; Yang, Y.; Chen, J.; Wu, Z.; Liu, Y.; Xie, S.X. Nitrifying activity and ammonia-oxidizing microorganisms in a constructed wetland treating polluted surface water. Sci. Total Environ. 2018, 628–629, 310–318. [Google Scholar] [CrossRef]
- Fan, L.F.; Chen, H.J.; Hsieh, H.L.; Lin, H.J.; Tang, S.L. Comparing abundance, composition and environmental influences on prokaryotic ammonia oxidizers in two subtropical constructed wetlands. Ecol. Eng. 2016, 90, 336–346. [Google Scholar] [CrossRef]
- Lee, J.A.; Francis, C.A. Spatiotemporal Characterization of San Francisco Bay Denitrifying Communities: A Comparison of nirK and nirS Diversity and Abundance. Microb. Ecol. 2017, 73, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Geets, J.; Cooman, M.D.; Wittebolle, L.; Heylen, K.; Vanparys, B.; Vos, P.D.; Verstraete, W.; Boon, N. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 2007, 75, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.Y.; Fan, F.L.; Yin, C.; Song, A.L.; Huang, P.R.; Tang, Y.J.; Zhu, P.; Peng, C.; Li, T.Q.; Wakelin, S.A.; et al. Long-term organic and inorganic fertilization alters temperature sensitivity of potential N2O emissions and associated microbes. Soil Biol. Biochem. 2016, 93, 131–141. [Google Scholar] [CrossRef]
- Baxter, A.M.; Johnson, L.; Edgerton, J.; Royer, T.; Leff, L.G. Structure and function of denitrifying bacterial assemblages in low-order Indiana streams. Freshw. Sci. 2012, 31, 304–317. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, C.L.; Ji, L.; Liu, Y.Q.; Xiao, J.; Cao, X.Y.; Zhou, Y.Y. Cause and effect of N/P ratio decline with eutrophication aggravation in shallow lakes. Sci. Total. Environ. 2018, 627, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Ding, L.J.; Xu, H.J.; Li, H.B.; Su, J.Q.; Zhu, Y.G. Variability in responses of bacterial communities and nitrogen oxide emission to urea fertilization among various flooded paddy soils. Fems Microbiol. Ecol. 2015, 91, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, H.H.; Dong, D.; Deng, H.; Strong, P.J.; Wang, H.L.; Wu, W.X. Insight into the Effects of Biochar on Manure Composting: Evidence Supporting the Relationship between N2O Emission and Denitrifying Community. Environ. Sci. Technol. 2013, 47, 7341–7349. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.E.; Buchwald, C.; McIlvin, M.R.; Casciotti, K.L. Isotopic signature of N2O produced by marine ammonia-oxidizing Archaea. Science 2011, 333, 1282–1285. [Google Scholar] [CrossRef]
- Cantera, J.J.; Stein, L.Y. Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 2007, 9, 765–776. [Google Scholar] [CrossRef]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.G.; Chan, P.P.; Gollabgir, A.; Hemp, J.; et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Song, C.L.; Cao, X.Y.; Zhou, Y.Y. The phosphorus release pathways and their mechanisms driven by organic carbon and nitrogen in sediments of eutrophic shallow lakes. Sci. Total. Environ. 2016, 572, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Hietanen, S.; Lukkari, K. Effects of short-term anoxia on benthic denitrification, nutrient fluxes and phosphorus forms in coastal Baltic sediment. Microb. Ecol. 2007, 49, 293–302. [Google Scholar] [CrossRef] [Green Version]
- Tao, H.; Feng, B.; Chen, W.; Hua, W.; Zhao, S.J. Behavior of dissolved organic nitrogen in sediment. Fresen. Environ. Bull. 2014, 23, 908–914. [Google Scholar]

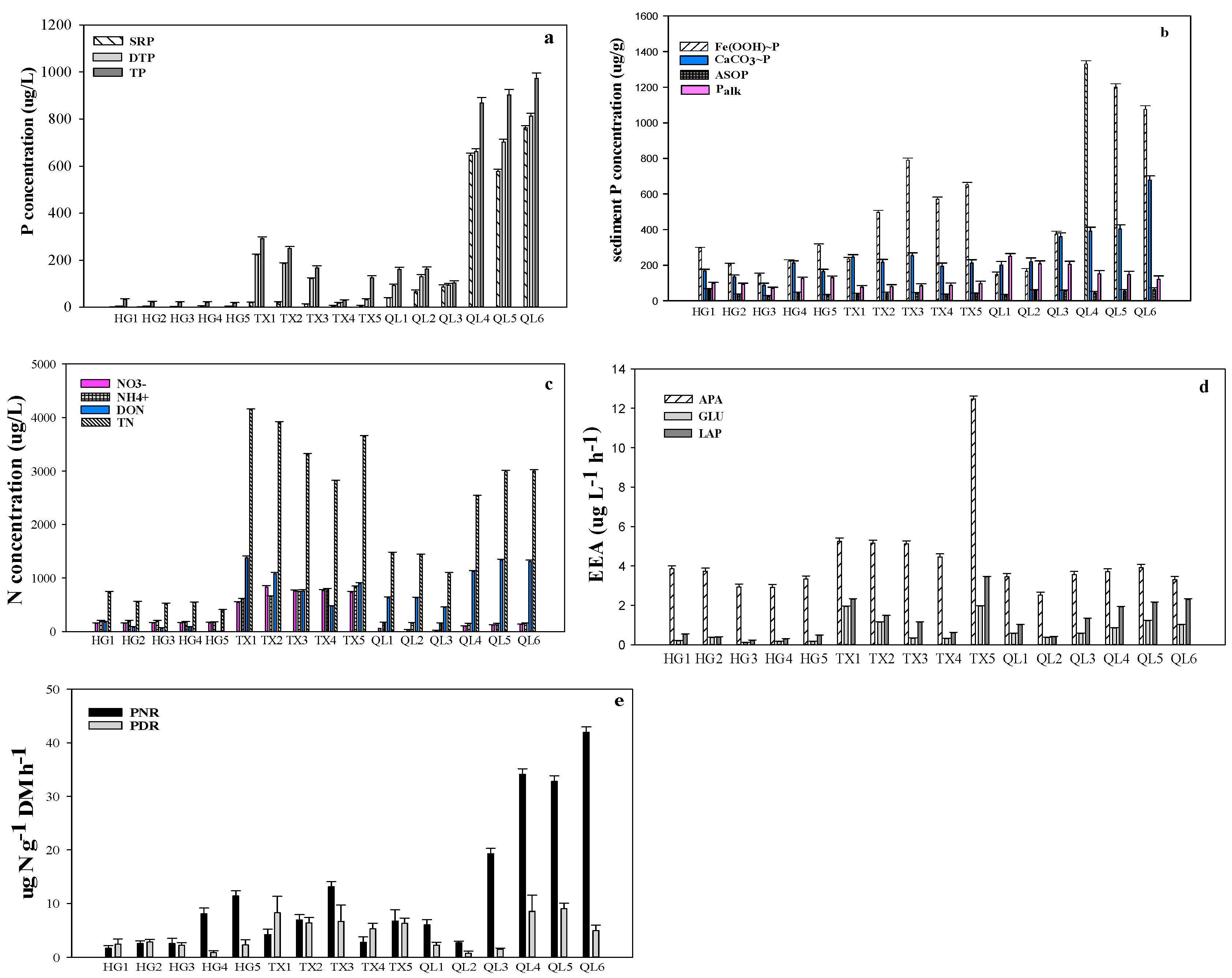


| Water Depth (m) | Transparency (m) | Chl a (μg L−1) | DO (mg L−1) | pH | TP (μg L−1) | TSI | |
|---|---|---|---|---|---|---|---|
| HG1 | 1.10 | 0.25 | 27.62 | 7.80 | 8.30 | 32.26 | 66.69 |
| HG2 | 1.20 | 0.30 | 20.41 | 7.90 | 8.30 | 21.98 | 63.41 |
| HG3 | 1.25 | 0.50 | 18.69 | 8.80 | 8.78 | 20.05 | 60.54 |
| HG4 | 1.30 | 0.65 | 16.92 | 8.30 | 8.67 | 20.69 | 58.96 |
| HG5 | 1.30 | 0.75 | 13.28 | 6.70 | 8.40 | 17.47 | 56.66 |
| TX1 | 2.30 | 0.25 | 487.36 | 15.24 | 9.83 | 290.74 | 87.07 |
| TX2 | 2.33 | 0.35 | 147.56 | 15.94 | 9.72 | 248.94 | 78.93 |
| TX3 | 2.41 | 0.35 | 92.07 | 14.26 | 9.72 | 167.29 | 75.50 |
| TX4 | 2.40 | 0.30 | 79.75 | 14.80 | 9.76 | 29.05 | 71.28 |
| TX5 | 2.43 | 0.20 | 194.45 | 17.70 | 9.96 | 124.21 | 81.16 |
| QL1 | 1.50 | 0.50 | 65.94 | 2.74 | 7.68 | 160.86 | 72.11 |
| QL2 | 1.50 | 0.45 | 48.48 | 5.74 | 7.90 | 162.79 | 70.96 |
| QL3 | 1.30 | 0.30 | 62.40 | 6.78 | 8.21 | 103.63 | 72.97 |
| QL4 | 1.50 | 0.30 | 222.64 | 8.50 | 8.70 | 868.76 | 84.71 |
| QL5 | 1.50 | 0.30 | 338.81 | 9.72 | 8.90 | 902.84 | 87.03 |
| QL6 | 1.50 | 0.30 | 296.47 | 9.75 | 8.93 | 972.92 | 86.50 |
| Title | AOA-amoA | AOB-amoA | nirK | nirS | nosZ1 | nosZ2 |
|---|---|---|---|---|---|---|
| HG1 | 1.11 × 107 | 1.89 × 107 | 2.27 × 106 | 1.02 × 107 | 1.46 × 108 | 1.24 × 108 |
| HG2 | 9.57 × 106 | 1.32 × 107 | 7.01 × 105 | 8.93 × 106 | 1.17 × 108 | 1.22 × 108 |
| HG3 | 1.67 × 107 | 2.00 × 108 | 1.25 × 106 | 1.14 × 107 | 4.38 × 108 | 3.09 × 108 |
| HG4 | 1.62 × 107 | 6.90 × 106 | 1.60 × 106 | 1.41 × 107 | 2.22 × 107 | 8.28 × 106 |
| HG5 | 1.04 × 107 | 9.46 × 106 | 2.67 × 106 | 9.62 × 106 | 4.45 × 107 | 1.24 × 107 |
| TX1 | 1.49 × 107 | 4.68 × 107 | 1.39 × 106 | 1.31 × 107 | 3.90 × 108 | 8.35 × 108 |
| TX2 | 1.11 × 107 | 2.31 × 107 | 3.18 × 106 | 1.02 × 107 | 4.27 × 108 | 3.88 × 108 |
| TX3 | 1.99 × 107 | 5.41 × 107 | 3.58 × 106 | 1.67 × 107 | 6.00 × 108 | 5.10 × 108 |
| TX4 | 3.09 × 107 | 2.67 × 108 | 3.43 × 107 | 2.45 × 107 | 3.63 × 108 | 7.48 × 108 |
| TX5 | 1.92 × 107 | 2.87 × 107 | 2.46 × 107 | 1.62 × 107 | 2.56 × 108 | 4.12 × 108 |
| QL1 | 4.83 × 107 | 6.68 × 108 | 2.03 × 106 | 3.56 × 107 | 5.85 × 108 | 3.64 × 108 |
| QL2 | 2.20 × 107 | 1.39 × 108 | 1.46 × 108 | 1.81 × 107 | 6.21 × 108 | 2.68 × 108 |
| QL3 | 4.52 × 107 | 2.75 × 108 | 1.48 × 108 | 3.36 × 107 | 1.74 × 108 | 7.65 × 108 |
| QL4 | 6.90 × 107 | 1.75 × 109 | 8.46 × 107 | 4.83 × 107 | 3.06 × 108 | 7.60 × 108 |
| QL5 | 5.15 × 107 | 1.94 × 109 | 2.82 × 107 | 3.78 × 107 | 9.44 × 108 | 7.30 × 108 |
| QL6 | 6.48 × 107 | 1.06 × 109 | 8.15 × 107 | 4.61 × 107 | 3.00 × 108 | 5.44 × 108 |
| AOA | AOB | nirK | nirS | nosZ1 | nosZ2 | PDR | PNR | |
|---|---|---|---|---|---|---|---|---|
| SRP | 0.845 ** | 0.795 ** | 0.675 ** | 0.844 ** | 0.598 * | 0.727 ** | 0.667 ** | 0.301 |
| DTP | 0.696 ** | 0.670 ** | 0.545 * | 0.695 ** | 0.644 ** | 0.653 ** | 0.674 ** | 0.546 * |
| Fe(OOH)~P | 0.546 * | 0.449 | 0.536 * | 0.548 * | 0.253 | 0.415 | 0.739 ** | 0.717 ** |
| Ca(OOH)~P | 0.780 ** | 0.574 * | 0.682 ** | 0.780 ** | 0.218 | 0.401 | 0.827 ** | 0.352 |
| ASOP | 0.311 | 0.230 | 0.562 * | 0.370 | 0.108 | 0.200 | 0.318 | −0.032 |
| Palk | 0.595 * | 0.427 | 0.492 | 0.592 * | 0.027 | −0.025 | 0.345 | −0.445 |
| APA | −0.237 | −0.199 | −0.010 | −0.116 | 0.169 | 0.317 | 0.004 | 0.604 * |
| GLU | 0.315 | 0.343 | 0.357 | 0.410 | 0.470 | 0.630 ** | 0.414 | 0.652 ** |
| LAP | 0.523 * | 0.405 | 0.422 | 0.524 * | 0.422 | 0.615 * | 0.604 | 0.726 ** |
| NO3−-N | −0.386 | −0.377 | −0.367 | −0.405 | 0.074 | 0.081 | −0.183 | 0.621 * |
| NO2−-N | −0.383 | −0.268 | −0.009 | −0.31 | 0.155 | 0.148 | −0.079 | 0.641 * |
| NH4+-N | −0.297 | −0.319 | −0.172 | −0.318 | 0.233 | 0.294 | −0.274 | 0.506 * |
| TN | 0.352 | 0.421 | 0.403 | 0.44 | 0.672 ** | 0.750 ** | 0.347 | 0.742 ** |
| TSI | 0.596 * | 0.588 * | 0.494 | 0.597 * | 0.660 ** | 0.795 ** | 0.501 * | 0.722 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Song, C.; Zhou, Z.; Cao, X.; Zhou, Y. Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes. Water 2019, 11, 1809. https://doi.org/10.3390/w11091809
Zhang Y, Song C, Zhou Z, Cao X, Zhou Y. Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes. Water. 2019; 11(9):1809. https://doi.org/10.3390/w11091809
Chicago/Turabian StyleZhang, Yao, Chunlei Song, Zijun Zhou, Xiuyun Cao, and Yiyong Zhou. 2019. "Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes" Water 11, no. 9: 1809. https://doi.org/10.3390/w11091809
APA StyleZhang, Y., Song, C., Zhou, Z., Cao, X., & Zhou, Y. (2019). Coupling between Nitrification and Denitrification as well as Its Effect on Phosphorus Release in Sediments of Chinese Shallow Lakes. Water, 11(9), 1809. https://doi.org/10.3390/w11091809




