Biodegradation of Amoxicillin, Tetracyclines and Sulfonamides in Wastewater Sludge
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
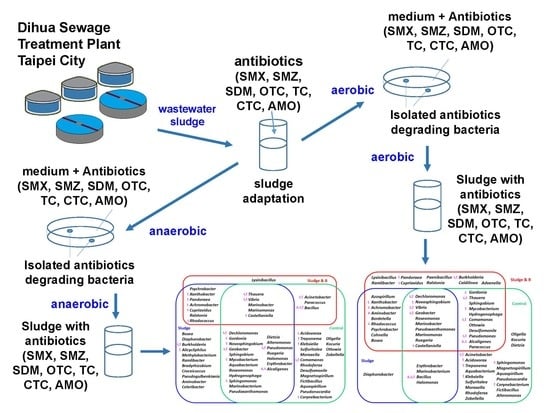
2.2. Sludge Sample and Medium
2.3. Sludge Adaptation
2.4. Enrichment, Isolation, and Identification of Antibiotic-Degrading Bacteria
2.5. Settings of Batch and Readdition Experiments
2.6. Analytical Methods
2.7. Microbial Community Analysis
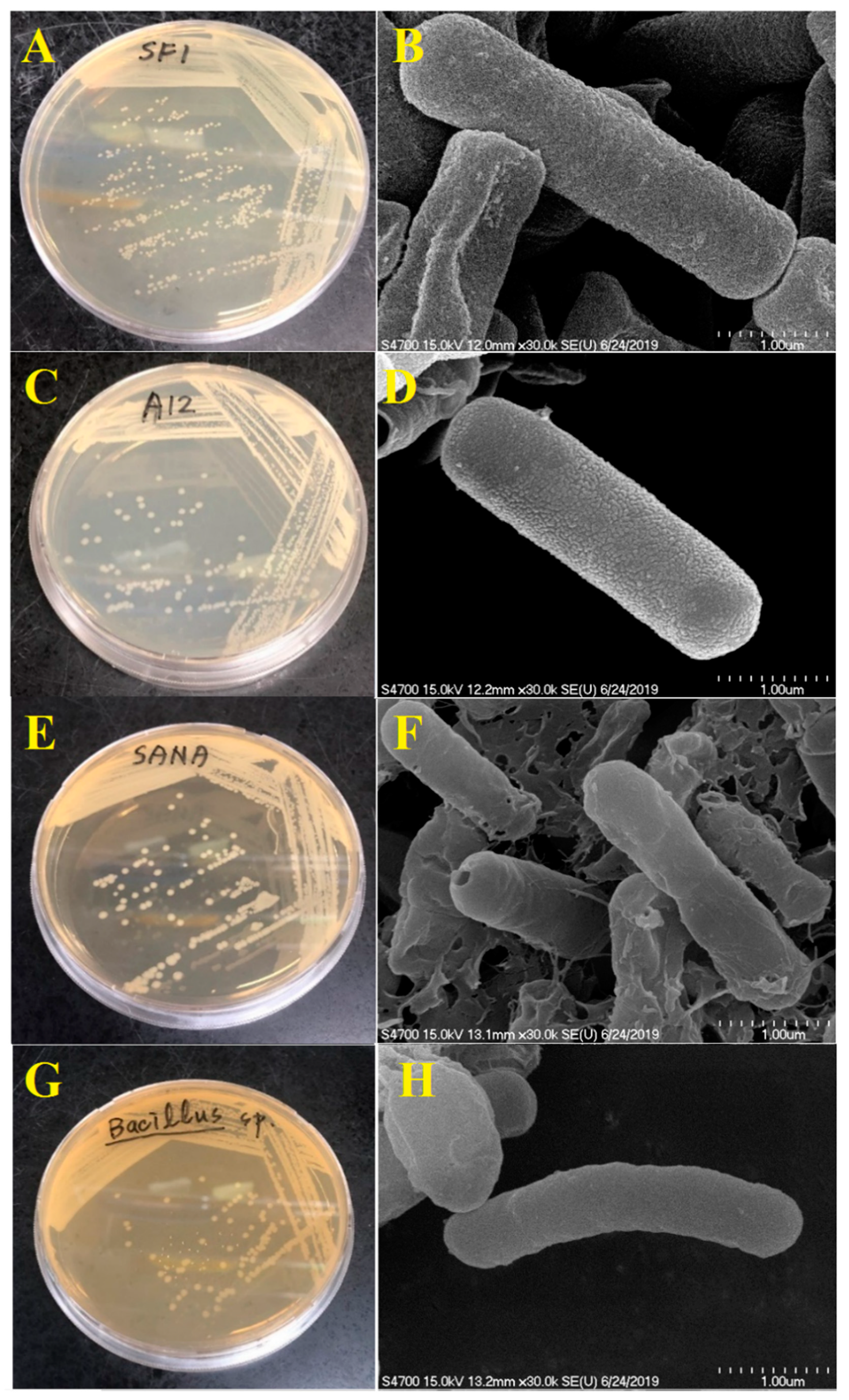
2.8. Scanning Electron Microscopy
3. Results and Discussion
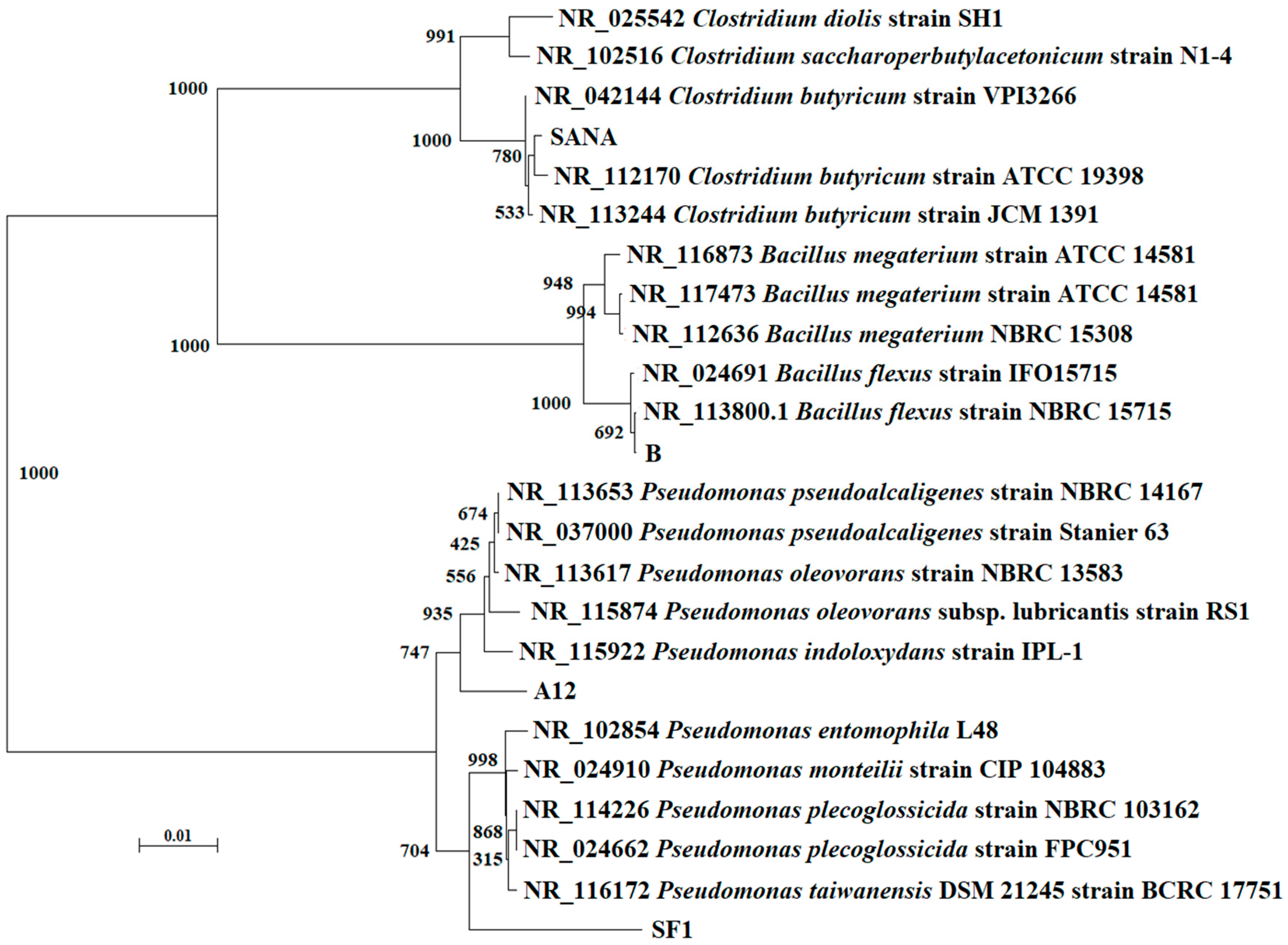
3.1. Isolation and Identification of Antibiotic-Degrading Bacteria
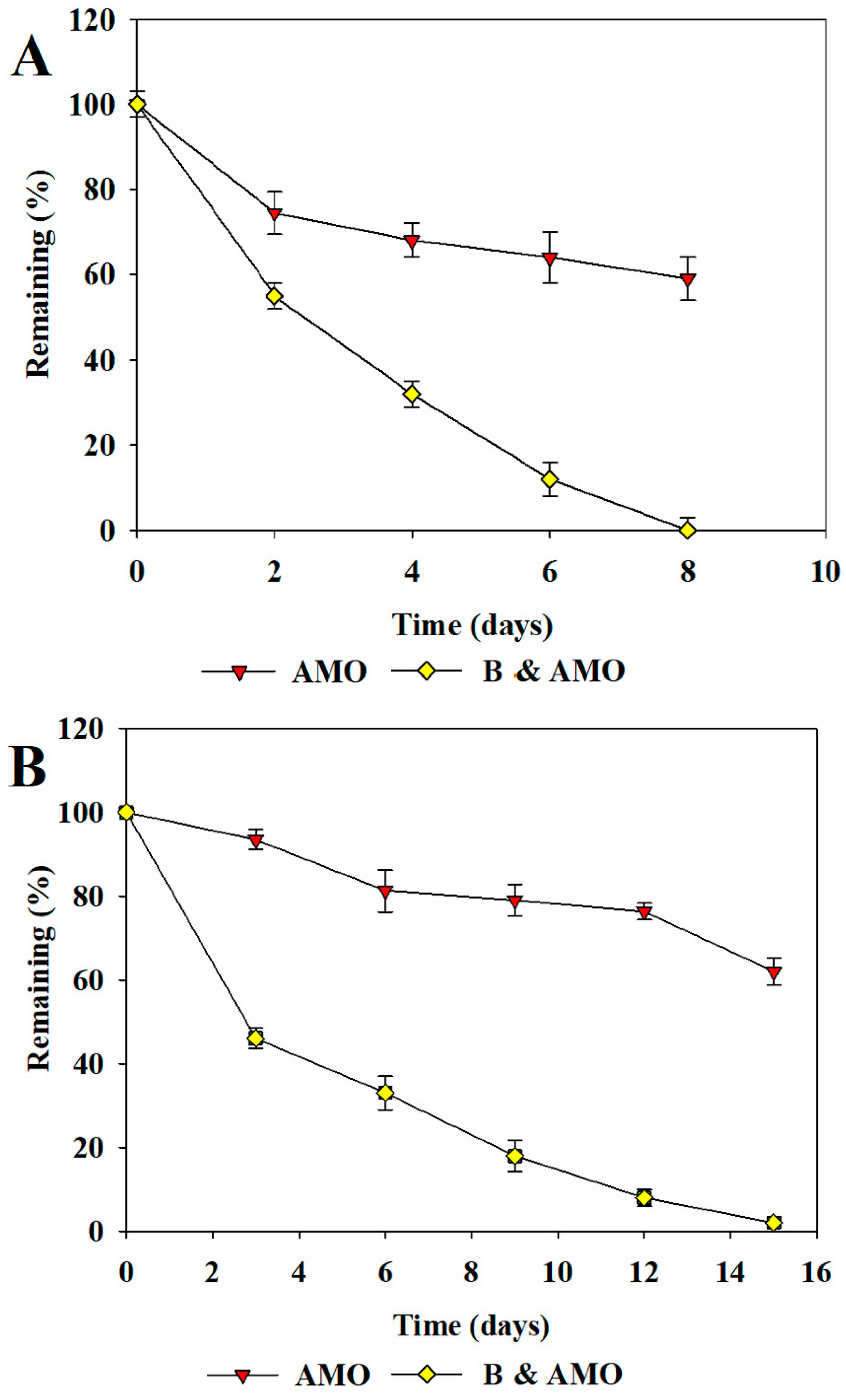
3.2. Degradation of Antibiotics in Sludge with Isolated Bacteria
3.3. Repeated Addition of Antibiotics and Their Degradation in Sludge
3.4. Microbial Communities in the Repeated Addition Experiments
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Salgado, R.; Noronha, J.P.; Oehmen, A.; Carvalho, G.; Reis, M.A. Analysis of 65 pharmaceuticals and personal care products in 5 wastewater treatment plants in Portugal using a simplified analytical methodology. Water Sci. Technol. 2010, 62, 2862–2871. [Google Scholar] [CrossRef]
- Barber, L.B.; Keefe, S.H.; LeBlanc, D.R.; Bradley, P.M.; Chapelle, F.H.; Meyer, M.T.; Loftin, K.A.; Kolpin, D.W.; Rubio, F. Fate of sulfamethoxazole, 4- nonylphenol, and 17ß-estradiol in groundwater contaminated by wastewater treatment plant effluent. Environ. Sci. Technol. 2009, 43, 4843–4850. [Google Scholar] [CrossRef] [Green Version]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Gzyl, K.E.; Wieden, H.J. Tetracycline does not directly inhibit the function of bacterial elongation factor Tu. PLoS ONE 2017, 12, e0178523. [Google Scholar] [CrossRef] [Green Version]
- Sukul, P.; Spiteller, M. Sulfonamides in the environment as veterinary drugs. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2006; Volume 187, pp. 67–101. [Google Scholar]
- Gobel, A.; Thomsen, A.; McArdell, C.S.; Joss, A.; Giger, W. Occurrence and sorption behavior of sulfonamides, macrolides, and trimethoprim in activated sludge treatment. Environ. Sci. Technol. 2005, 39, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Nieto, A.; Borrull, F.; Pocurull, E.; Marce, R.M. Occurrence of pharmaceuticals and hormones in sewage sludge. Environ. Toxicol. Chem. 2010, 29, 1484–1489. [Google Scholar] [CrossRef]
- Adamek, E.; Baran, W.; Sobczak, A. Assessment of the biodegradability of selected sulfa drugs in two polluted rivers in Poland: Effects of seasonal variations, accidental contamination, turbidity and salinity. J. Hazard Mater. 2016, 313, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Baghapour, M.A.; Shirdarreh, M.R.; Faramarzian, M. Amoxicillin removal from aqueous solutions using submerged biological aerated filter. Desalin. Water Treat. 2014, 54, 790–801. [Google Scholar] [CrossRef]
- Gavrilescu, M.; Demnerova, K.; Aamand, J.; Agathos, S.; Fava, F. Emerging pollutants in the environment: Present and future challenges in biomonitoring, ecological risks and bioremediation. New Biotechnol. 2015, 32, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. The role of operating parameters and oxidative damage mechanisms of advanced chemical oxidation processes in the combat against antibiotic-resistant bacteria and resistance genes present in urban wastewater. Water Res. 2018, 129, 208–230. [Google Scholar] [PubMed]
- Chang, B.V.; Fan, S.N.; Tsai, Y.C.; Chung, Y.L.; Tu, P.X.; Yang, C.W. Removal of emerging contaminants using spent mushroom compost. Sci. Total Environ. 2018, 634, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, W.; Chen, Z.; Qin, W.; Wen, X. Responses of antibiotics, antibiotic resistance genes, and mobile genetic elements in sewage sludge to thermal hydrolysis pre-treatment and various anaerobic digestion conditions. Environ. Int. 2019, 133, 105156. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Wei, Z.; Zhao, Y.; Zhang, D.; Pan, X. Enhanced performance of tetracycline treatment in wastewater using aerobic granular sludge with in-situ generated biogenic manganese oxides. Sci. Total Environ. 2020, 735, 139533. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Yang, Q.; Li, X.; Yuan, W.; Chen, Y.; Wang, R. Impacts of long-term exposure to tetracycline and sulfamethoxazole on the sludge granules in an anoxic-aerobic wastewater treatment system. Sci. Total Environ. 2019, 684, 67–77. [Google Scholar] [CrossRef]
- Krzeminski, P.; Tomei, M.C.; Karaolia, P.; Langenhoff, A.; Almeida, C.M.; Felis, E.; Gritten, F.; Andersen, H.R.; Fernandes, T.; Manaia, C.M.; et al. Performance of secondary wastewater treatment methods for the removal of contaminants of emerging concern implicated in crop uptake and antibiotic resistance spread: A review. Sci. Total Environ. 2019, 648, 1052–1081. [Google Scholar] [CrossRef] [Green Version]
- Bisognin, R.P.; Wolff, D.B.; Carissimi, E.; Prestes, O.D.; Zanella, R. Occurrence and fate of pharmaceuticals in effluent and sludge from a wastewater treatment plant in Brazil. Environ. Technol. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Ezzariai, A.; Hafidi, M.; Khadra, A.; Aemig, Q.; El Fels, L.; Barret, M.; Merlina, G.; Patureau, D.; Pinelli, E. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. J. Hazard Mater. 2018, 359, 465–481. [Google Scholar] [CrossRef]
- Boonnorat, J.; Kanyatrakul, A.; Prakhongsak, A.; Honda, R.; Panichnumsin, P.; Boonapatcharoen, N. Effect of hydraulic retention time on micropollutant biodegradation in activated sludge system augmented with acclimatized sludge treating low-micropollutants wastewater. Chemosphere 2019, 230, 606–615. [Google Scholar] [CrossRef]
- Nas, B.; Argun, M.E.; Dolu, T.; Ateş, H.; Yel, E.; Koyuncu, S.; Dinç, S.; Kara, M. Occurrence, loadings and removal of EU-priority polycyclic aromatic hydrocarbons (PAHs) in wastewater and sludge by advanced biological treatment, stabilization pond and constructed wetland. J. Environ. Manag. 2020, 268, 110580. [Google Scholar] [CrossRef]
- Kamaz, M.; Wickramasinghe, S.R.; Eswaranandam, S.; Zhang, W.; Jones, S.M.; Watts, M.J.; Qian, X. Investigation into micropollutant removal from wastewaters by a membrane bioreactor. Int. J. Environ. Res. Public Health 2019, 16, 1363. [Google Scholar] [CrossRef] [Green Version]
- Gauthier, H.; Yargeau, V.; Cooper, D.G. Biodegradation of pharmaceuticals by Rhodococcus rhodochrous and Aspergillus niger by co-metabolism. Sci. Total Environ. 2010, 408, 1701–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, Y.; Bao, J.; Chang, G.; Zheng, H.; Li, X.; Du, J.; Snow, D. Biotransformation of tetracycline by a novel bacterial strain Stenotrophomonas maltophilia DT1. J. Hazard. Mater. 2016, 318, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, S.; Hu, Y.; Cheng, J.; Chen, Y. Biodegradation mechanism of tetracycline (TEC) by strain Klebsiella sp. SQY5 as revealed through products analysis and genomics. Ecotoxicol. Environ. Saf. 2019, 85, 109676–109683. [Google Scholar] [CrossRef]
- Larcher, S.; Yargeau, V. Biodegradation of sulfamethoxazole by individual and mixed bacteria. Appl. Microbiol. Biotechnol. 2011, 91, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S. Microbial degradation of sulfamethoxazole in the environment. Appl. Microbiol. Biotechnol. 2018, 102, 3573–3582. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, B.; Zou, R.; Xie, S.; Yuan, B. Antibiotic sulfanilamide biodegradation by acclimated microbial populations. Appl. Microbiol. Biotechnol. 2016, 100, 2439–2447. [Google Scholar] [CrossRef]
- Liao, X.; Zou, R.; Li, B.; Tong, T.; Xie, S.; Yuan, B. Biodegradation of chlortetracycline by acclimated microbiota. Process Saf. Environ. 2017, 109, 11–17. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Chang, B.V.; Chang, Y.T.; Chao, W.L.; Yeh, S.L.; Kuo, D.L.; Yang, C.W. Effects of sulfamethoxazole and sulfamethoxazole-degrading bacteria on water quality and microbial communities in milkfish ponds. Environ. Pollut. 2019, 252, 305–316. [Google Scholar] [CrossRef]
- Yang, C.W.; Chao, W.L.; Hsieh, C.Y.; Chang, B.V. Biodegradation of malachite green in milkfish pond sediments. Sustainability 2019, 11, 4179. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.T.; Tai, C.J.; Wu, Y.C.; Chen, Y.B.; Lee, F.L.; Wang, S.L. Pseudomonas taiwanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Micr. 2010, 60, 2094–2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huertas, M.J.; Luque-Almagro, V.M.; Martinez-Luque, M.; Blasco, R.; Moreno-Vivian, C.; Castillo, F.; Roldan, M.D. Cyanide metabolism of Pseudomonas pseudoalcaligenes CECT5344: Role of siderophores. Biochem. Soc. Trans. 2006, 34, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. New Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Ekambaram, S.P.; Perumal, S.S.; Annamalai, U. Decolorization and biodegradation of remazol reactive dyes by Clostridium species. 3 Biotech. 2016, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.W.; Hsiao, W.C.; Chang, B.V. Biodegradation of sulfonamide antibiotics in sludge. Chemosphere 2016, 150, 559–565. [Google Scholar] [CrossRef]
- Suda, T.; Hata, T.; Kawai, S.; Okamura, H.; Nishida, T. Treatment of tetracycline antibiotics by laccase in the presence of 1-hydroxybenzotriazole. Bioresour. Technol. 2012, 103, 498–501. [Google Scholar] [CrossRef] [Green Version]
- Pepper, L.L.; Gerba, C.P.; Gentry, T.J. Environmental microbiology. In Microorganisms and Organic Pollutant; Maier, M.M., Gentry, T.J., Eds.; Elsevier Inc.: Berkeley, CA, USA, 2015; pp. 377–413. [Google Scholar]
- Yin, Z.; Xia, D.; Shen, M.; Zhu, D.; Cai, H.; Wu, M.; Zhu, Q.; Kang, Y. Tetracycline degradation by Klebsiella sp. strain TR5: Proposed degradation pathway and possible genes involved. Chemosphere 2020, 253, 126729. [Google Scholar] [CrossRef]
- Wu, X.; Gu, Y.; Wu, X.; Zhou, X.; Zhou, H.; Amanze, C.; Shen, L.; Zeng, W. Construction of a Tetracycline Degrading Bacterial Consortium and Its Application Evaluation in Laboratory-Scale Soil Remediation. Microorganisms 2020, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.; Huang, J.; Cao, J.; Xu, J.; Mi, J.; Wang, Y.; Ma, B.; Zou, Y.; Liao, X.; Liang, J.B.; et al. Heterologous expression of the tetracycline resistance gene tetX to enhance degradability and safety in doxycycline degradation. Ecotoxicol. Environ. Saf. 2020, 191, 110214. [Google Scholar] [CrossRef]
- Sodhi, K.K.; Kumar, M.; Singh, D.K. Potential application in amoxicillin removal of Alcaligenes sp. MMA and enzymatic studies through molecular docking. Arch. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.H.; Hu, Y. Simultaneous sulfamethoxazole biodegradation and nitrogen conversion by Achromobacter sp. JL9 using with different carbon and nitrogen sources. Bioresour. Technol. 2019, 293, 122061. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.Y.; Silva, A.F.; Reis, A.C.; Nunes, O.C.; Rodrigues, A.M.; Rodrigues, J.E.; Cardoso, V.V.; Benoliel, M.J.; Reis, M.A.; Oehmen, A.; et al. Bioaugmentation of membrane bioreactor with Achromobacter denitrificans strain PR1 for enhanced sulfamethoxazole removal in wastewater. Sci. Total Environ. 2019, 648, 44–55. [Google Scholar] [CrossRef]
- Wang, S.; Hu, Y.; Wang, J. Biodegradation of typical pharmaceutical compounds by a novel strain Acinetobacter sp. J. Environ. Manag. 2018, 217, 240–246. [Google Scholar] [CrossRef] [PubMed]


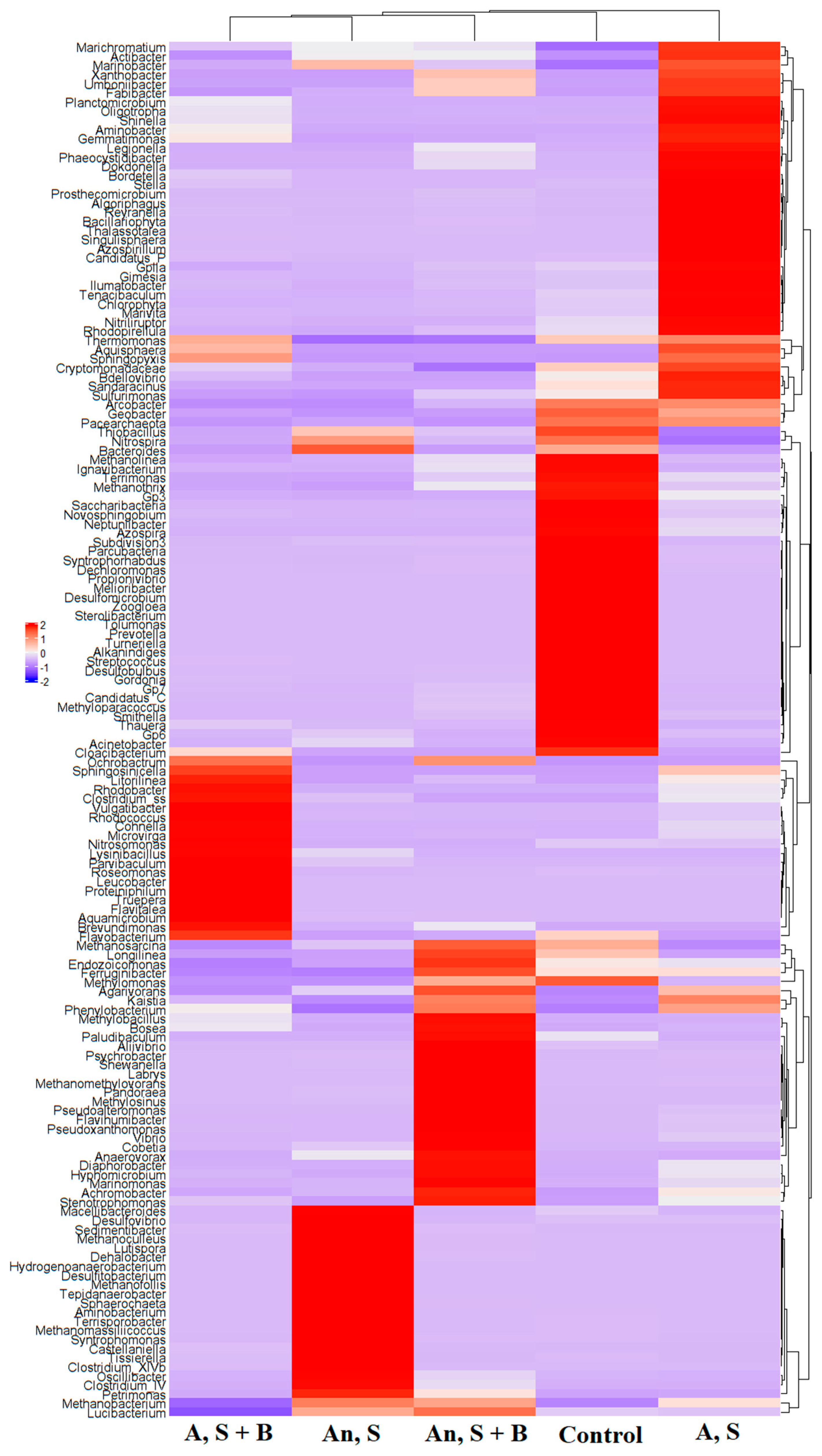
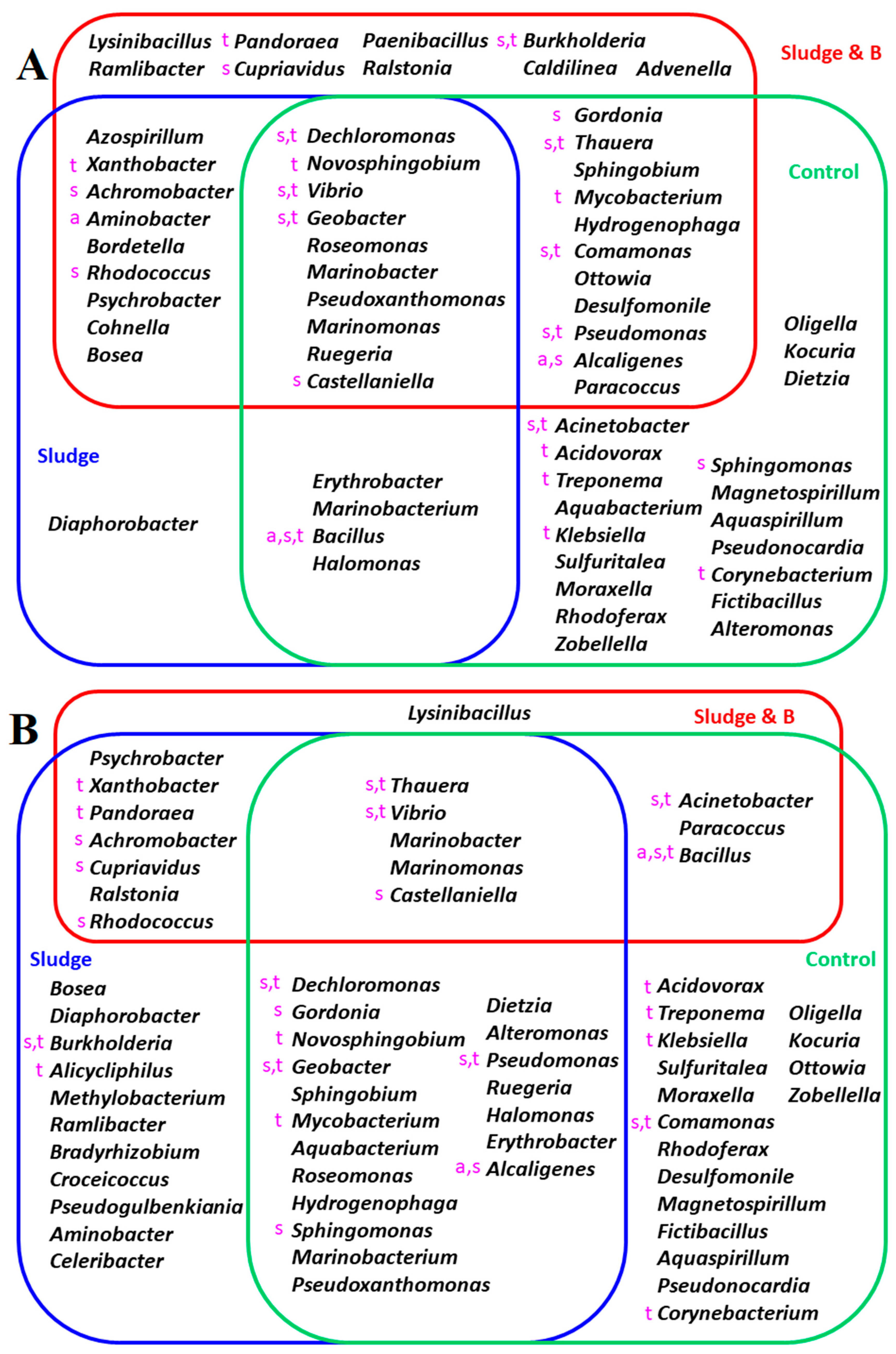

| Aerobic Condition a | Anaerobic Condition b | |||||||
|---|---|---|---|---|---|---|---|---|
| Medium | SF1 | A12 | SF1+A12 | Medium | B | SANA | B + SANA | |
| CTC | 97.2 ± 3.70 | 18.4 ± 0.93 | 30.6 ± 1.48 | 10.6 ± 0.53 | 98.3 ± 4.33 | 18.4 ± 0.94 | 40.6 ± 2.13 | 10.4 ± 0.42 |
| AMO | 94.5 ± 2.91 | 10.9 ± 0.49 | 18.6 ± 0.88 | 6.6 ± 0.34 | 96.7 ± 5.24 | 10.9 ± 0.58 | 18.6 ± 0.90 | 6.4 ± 0.29 |
| SMX | 98.6 ± 5.12 | 4.1 ± 0.12 | 10.4 ± 0.41 | 0.7 ± 0.02 | 99.1 ± 6.78 | 4.9 ± 0.10 | 10.4 ± 0.44 | 0.5 ± 0.01 |
| CTC | AMO | SMX | ||
|---|---|---|---|---|
| 1st | Sludge a | 35.6 ± 1.6 | 41.6 ± 1.8 | 46.3 ± 1.5 |
| Sludge & SF1 & A12 a | 82.6 ± 2.5 | 88.2 ± 2.9 | 90.6 ± 3.8 | |
| Sludge b | 33.5 ± 1.2 | 37.4 ± 1.1 | 42.2 ± 1.6 | |
| Sludge & B & SANAb | 85.2 ± 2.4 | 88.1 ± 2.1 | 92.3 ± 3.1 | |
| 2nd | Sludge a | 43.7 ± 1.5 | 45.3 ± 1.3 | 49.1 ± 1.4 |
| Sludge & SF1 & A12 a | 88.6 ± 2.5 | 90.2 ± 2.9 | 92.5 ± 2.9 | |
| Sludge b | 41.2 ± 1.5 | 43.7 ± 1.3 | 46.4 ± 1.2 | |
| Sludge & B & SANA b | 90.7 ± 3.3 | 91.1 ± 3.1 | 94.3 ± 3.5 | |
| 3rd | Sludge a | 46.1 ± 1.3 | 50.4 ± 1.3 | 52.5 ± 1.1 |
| Sludge & SF1 & A12 a | 90.3 ± 3.2 | 95.3 ± 4.1 | 97.5 ± 3.9 | |
| Sludge b | 42.7 ± 1.1 | 46.4 ± 1.3 | 50.4 ± 1.2 | |
| Sludge & B & SANA b | 92.4 ± 4.2 | 96.4 ± 4.4 | 98.1 ± 4.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-W.; Liu, C.; Chang, B.-V. Biodegradation of Amoxicillin, Tetracyclines and Sulfonamides in Wastewater Sludge. Water 2020, 12, 2147. https://doi.org/10.3390/w12082147
Yang C-W, Liu C, Chang B-V. Biodegradation of Amoxicillin, Tetracyclines and Sulfonamides in Wastewater Sludge. Water. 2020; 12(8):2147. https://doi.org/10.3390/w12082147
Chicago/Turabian StyleYang, Chu-Wen, Chien Liu, and Bea-Ven Chang. 2020. "Biodegradation of Amoxicillin, Tetracyclines and Sulfonamides in Wastewater Sludge" Water 12, no. 8: 2147. https://doi.org/10.3390/w12082147