Cell-Free Synthesis Meets Antibody Production: A Review
Abstract
:1. Introduction
2. Conventional Antibody Production Technologies
3. Cell-Free Protein Synthesis
3.1. Disulfide Bond Formation in Cell-Free Systems
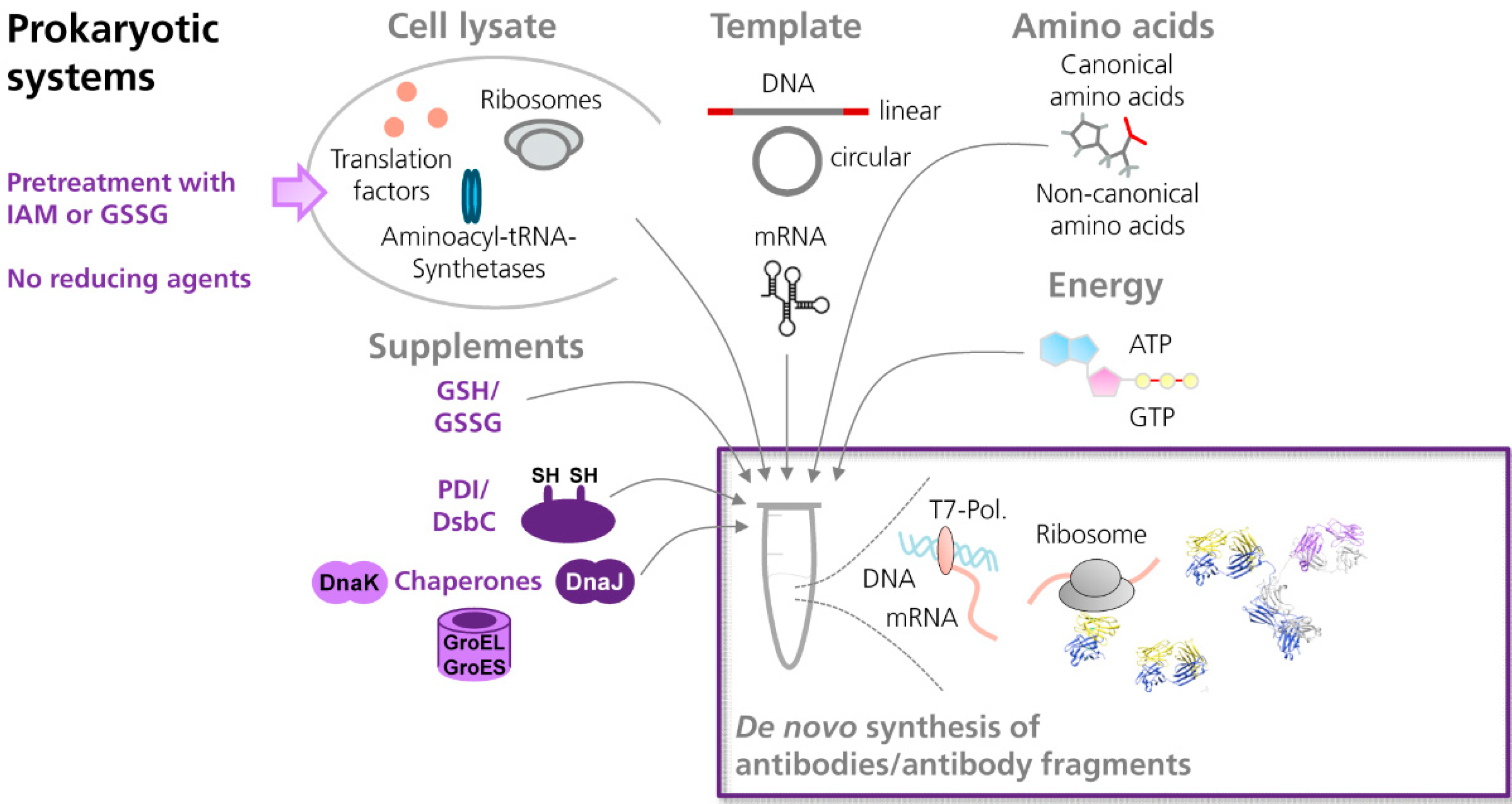
3.2. Production of Antibodies in Cell-Free Systems
3.2.1. Prokaryotic Cell-Free Systems
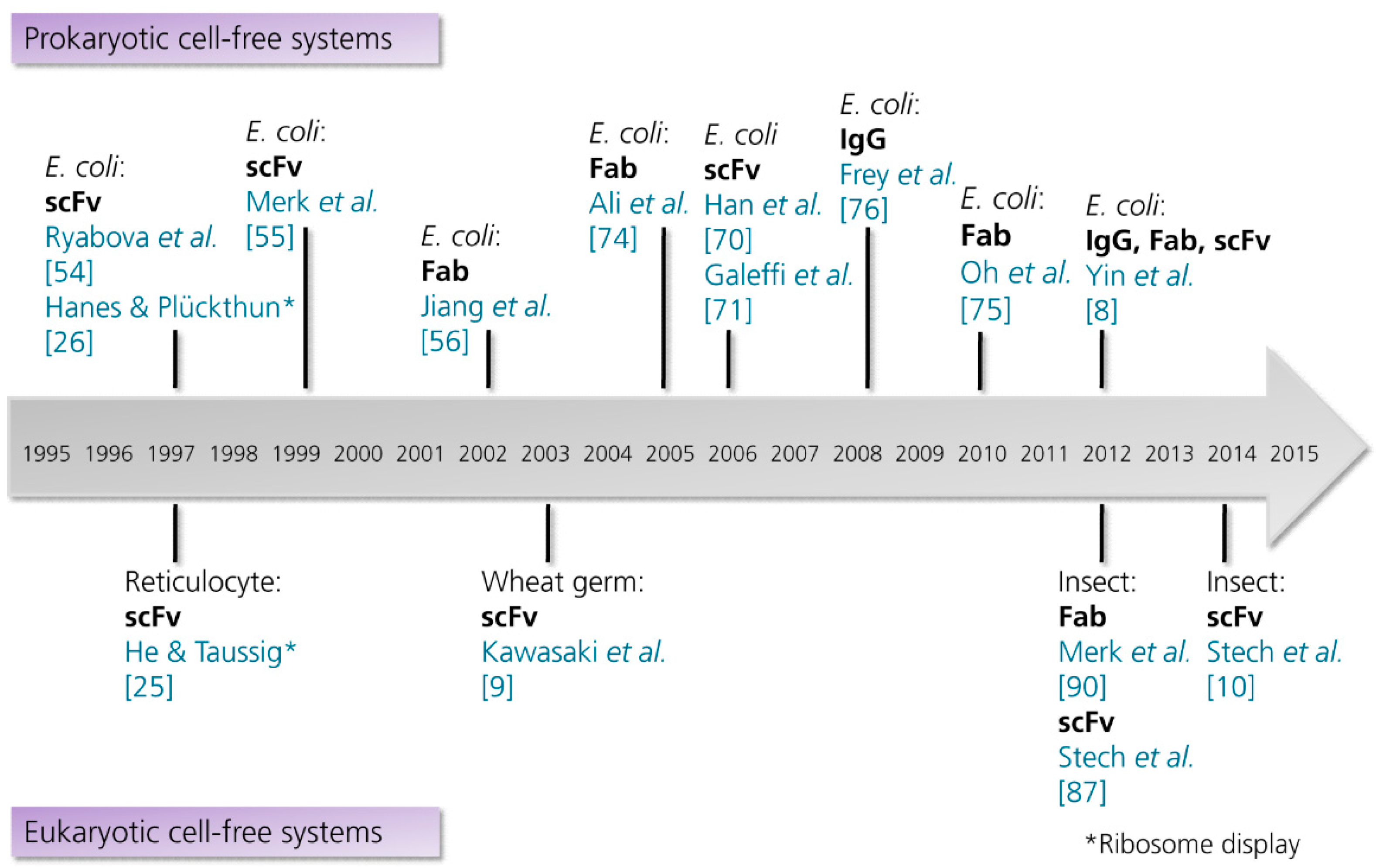
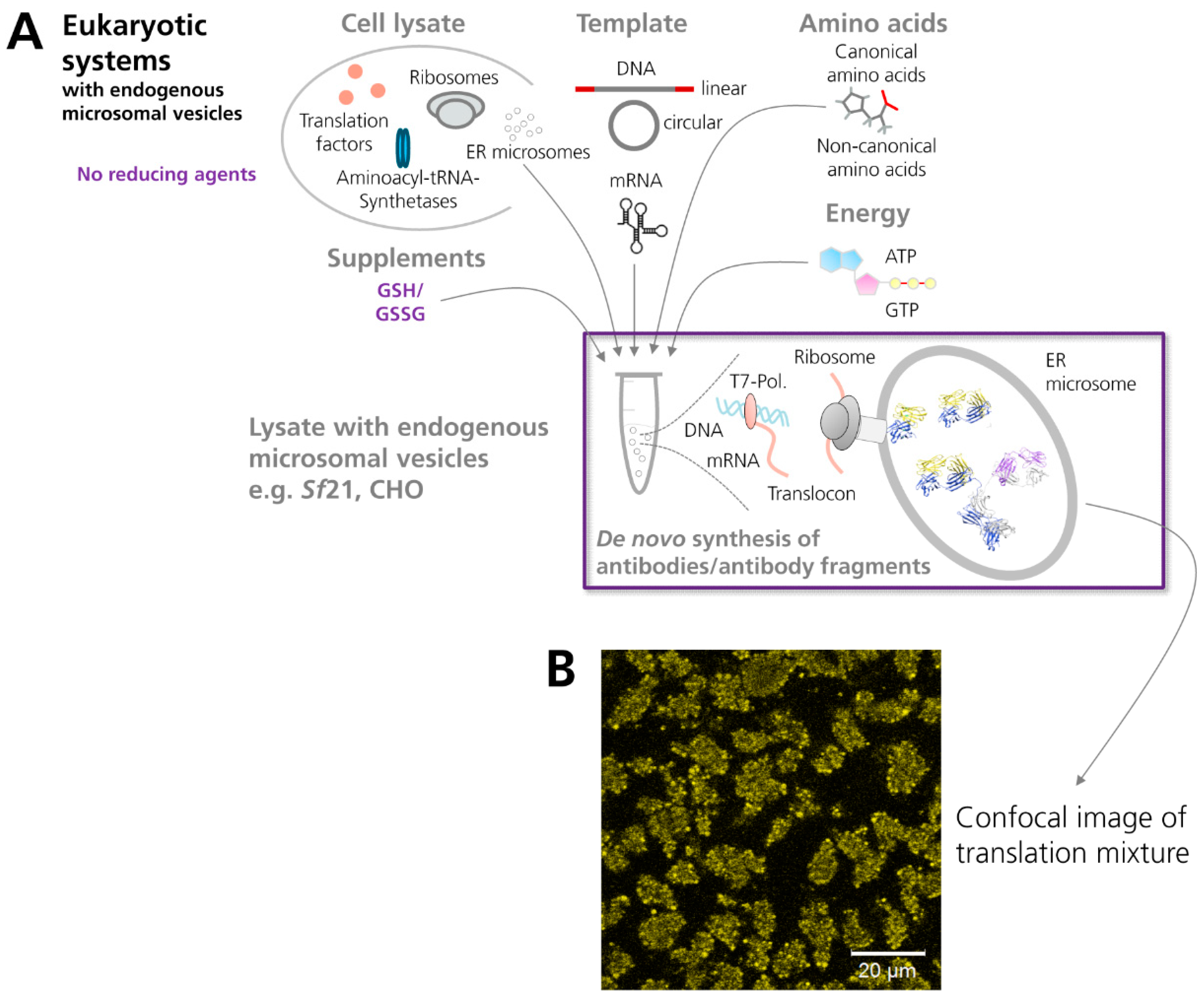
3.2.2. Eukaryotic Cell-Free Systems
3.2.3. Cell-Free Systems as a Source of Modified Antibodies and Antibody Fragments
3.2.3.1. scFv Fusion Proteins
3.2.3.2. Labeling of Antibodies and Antibody Fragments with Non-canonical Amino Acids
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hudson, P.; Souriau, C. Engineered antibodies. Nat. Med. 2003, 9, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Hagemeyer, C.E.; von zur Muhlen, C.; von Elverfeldt, D.; Peter, K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb. Haemost. 2009, 101, 1012–1019. [Google Scholar] [PubMed]
- Goshima, N.; Kawamura, Y.; Fukumoto, A.; Miura, A.; Honma, R.; Satoh, R.; Wakamatsu, A.; Yamamoto, J.; Kimura, K.; Nishikawa, T.; et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 2008, 5, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M.; Gold, D.S.; Heinsohn, H.G.; Murray, C.J. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Brödel, A.K.; Quast, R.B.; Sachse, R.; Kubick, S. Cell-free systems: Functional modules for synthetic and chemical biology. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 67–102. [Google Scholar]
- Orth, J.H.C.; Schorcha, B.; Boundy, S.; Ffrench-Constant, R.; Kubick, S.; Aktories, K. Cell-free synthesis and characterization of a novel cytotoxic pierisin-like protein from the cabbage butterfly Pieris rapae. Toxicon 2011, 57, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Katzen, F.; Peterson, T.C.; Kudlicki, W. Membrane protein expression: No cells required. Trends Biotechnol. 2009, 27, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Garces, E.D.; Yang, J.; Zhang, J.; Tran, C.; Steiner, A.R.; Roos, C.; Bajad, S.; Hudak, S.; Penta, K.; et al. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs 2012, 4, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Gouda, M.D.; Sawasaki, T.; Takai, K.; Endo, Y. Efficient synthesis of a disulfide-containing protein through a batch cell-free system from wheat germ. Eur. J. Biochem. 2003, 270, 4780–4786. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Hust, M.; Schulze, C.; Dübel, S.; Kubick, S. Cell-free eukaryotic systems for the production, engineering and modification of scFv antibody fragments. Eng. Life Sci. 2014, 14, 387–398. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Presta, L.G. Engineering of therapeutic antibodies to minimize immunogenicity and optimize function. Adv. Drug Deliver. Rev. 2006, 58, 640–656. [Google Scholar] [CrossRef]
- Weisser, N.E.; Hall, J.C. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol. Adv. 2009, 27, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotný, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 242, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Mccafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domain. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Smith, G. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Barbas, C.F., 3rd; Kang, A.S.; Lerner, R.A.; Benkovic, S.J. Assembly of combinatorial antibody libraries on phages surfaces: The gene III site. Proc. Natl. Acad. Sci. USA 1991, 88, 7987–7982. [Google Scholar] [CrossRef]
- Barbas, C.; Lerner, R. Combinatorial immunoglobulin libraries on the surface of phage (Phabs): Rapid selection of antigen-specific Fabs. Methods 1991, 2, 119–124. [Google Scholar] [CrossRef]
- Hust, M.; Dübel, S. Mating antibody phage display to proteomics. Trends Biotechnol. 2004, 22, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Francisco, J.A.; Campbell, R.; Iverson, B.L.; Georgiou, G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc. Natl. Acad. Sci. USA 1993, 90, 10444–10448. [Google Scholar] [CrossRef] [PubMed]
- Boder, E.T.; Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997, 15, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.; Szostak, J. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 12297–12302. [Google Scholar] [CrossRef]
- Nemoto, N.; Miyamoto-Sato, E.; Husimi, Y.; Yanagawa, H. In vitro virus: Bonding of mRNA bearing puromycin at the 3'-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997, 414, 405–408. [Google Scholar] [CrossRef]
- He, M.; Taussig, M.J. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 1997, 25, 5132–5134. [Google Scholar] [CrossRef] [PubMed]
- Hanes, J.; Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 1997, 94, 4937–4942. [Google Scholar]
- Better, M.; Chang, C.P.; Robinson, R.R.; Horwitz, A.H. Escherichia coli secretion of an active chimeric antibody fragment. Science 1988, 240, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A.; Plückthun, A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 1988, 240, 1038–1041. [Google Scholar] [CrossRef] [PubMed]
- Carter, P.; Kelley, R.F.; Rodrigues, M.L.; Snedecor, B.; Covarrubias, M.; Velligan, M.D.; Wong, W.L.T.; Rowland, A.M.; Kotts, C.E.; Carver, M.E.; et al. High Level Escherichia coli Expression and Production of a Bivalent Humanized Antibody Fragment. Nat. Biotechnol. 1992, 10, 163–167. [Google Scholar] [CrossRef]
- Simmons, L.C.; Reilly, D.; Klimowski, L.; Shantha Raju, T.; Meng, G.; Sims, P.; Hong, K.; Shields, R.L.; Damicof, L.A.; Rancatoreg, P.; et al. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J. Immunol. Methods 2002, 263, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Rojas, G.; Mitchell, J.N.; Vincent, K.J.; Wu, J.; McCafferty, J.; Schofield, D.J. A simple vector system to improve performance and utilization of recombinant antibodies. BMC Biotechnol. 2006, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Hackel, B.; Huang, D.; Bubolz, J.C.; Wang, X.X.; Shusta, E.V. Production of soluble and active transferrin receptor-targeting single-chain antibody using Saccharomyces cerevisiae. Pharm. Res. 2006, 23, 790–797. [Google Scholar] [CrossRef]
- Ren, F.; Li, B.-C.; Zhang, N.-N.; Cao, M.; Dan, W.-B.; Zhang, S.-Q. Expression, purification and characterization of anti-BAFF antibody secreted from the yeast Pichia pastoris. Biotechnol. Lett. 2008, 30, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Bruenke, J.; Fischer, B.; Barbin, K.; Schreiter, K.; Wachter, Y.; Mahr, K.; Titgemeyer, F.; Niederweis, M.; Peipp, M.; Zunino, S.J.; et al. A recombinant bispecific single-chain Fv antibody against HLA class II and FcγRIII (CD16) triggers effective lysis of lymphoma cells. Br. J. Haematol. 2004, 125, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, S.P.; Franklin, S.E.; Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Makvandi-Nejad, S.; McLean, M.D.; Hirama, T.; Almquist, K.C.; MacKenzie, C.R.; Hall, J.C. Transgenic tobacco plants expressing a dimeric single-chain variable fragment (scfv) antibody against Salmonella enterica serotype Paratyphi B. Transgenic. Res. 2005, 14, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Natsume, A.; Wakitani, M.; Yamane-Ohnuki, N.; Shoji-Hosaka, E.; Niwa, R.; Uchida, K.; Satoh, M.; Shitara, K. Fucose removal from complex-type oligosaccharide enhances the antibody-dependent cellular cytotoxicity of single-gene-encoded bispecific antibody comprising of two single-chain antibodies linked to the antibody constant region. J. Biochem. 2006, 140, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.; James, D. Production of recombinant monoclonal antibodies in mammalian cells. In Cell Engineering; Al-Rubeai, M., Ed.; Springer: New York, NY, USA, 2009; Volume 6. [Google Scholar]
- Li, F.; Shen, A.; Amanullah, A. Cell culture processes for monoclonal antibody production. Pharm. Sci. 2013, 1–38. [Google Scholar]
- Wurm, F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004, 22, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug Deliver. Rev. 2006, 58, 671–685. [Google Scholar] [CrossRef]
- Pedersen, A.; Hellberg, K.; Enberg, J.; Karlsson, B.G. Rational improvement of cell-free protein synthesis. New Biotechnol. 2010, 28, 218–224. [Google Scholar] [CrossRef]
- Nirenberg, M.; Matthaei, J. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Bechlars, S.; Wüstenhagen, D.A.; Drägert, K.; Dieckmann, R.; Strauch, E.; Kubick, S. Cell-free synthesis of functional thermostable direct hemolysins of Vibrio parahaemolyticus. Toxicon 2013, 76, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.D.; Tawfik, D.S. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003, 22, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Klammt, C.; Koglin, A.; Löhr, F.; Schneider, B.; Dötsch, V.; Bernhard, F. Preparative scale cell-free expression systems: New tools for the large scale preparation of integral membrane proteins for functional and structural studies. Methods 2007, 41, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.; Padlan, E.; Sheriff, S. Antibody-antigen complexes. Annu. Rev. Biochem. 1990, 59, 439–473. [Google Scholar] [CrossRef] [PubMed]
- Glockshuber, R.; Schmidt, T.; Plückthun, A. The disulfide bonds in antibody variable domains: Effects on stability, folding in vitro, and functional expression in Escherichia coli. Biochemistry 1992, 31, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Hamaguchi, K. The role of the intrachain disulfide bond in the conformation and stability of the constant fragment of the immunoglobulin light chain. J. Biochem. 1979, 86, 1433–1441. [Google Scholar] [PubMed]
- Kim, D.-M.; Swartz, J.R. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004, 85, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Bundy, B.C.; Swartz, J.R. Efficient disulfide bond formation in virus-like particles. J. Biotechnol. 2011, 154, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Katzen, F.; Chang, G.; Kudlicki, W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005, 23, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Ryabova, L.A.; Desplancq, D.; Spirin, A.S.; Plückthun, A. Functional antibody production using cell-free translation: Effects of protein disulfide isomerase and chaperones. Nat. Biotechnol. 1997, 15, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Merk, H.; Stiege, W.; Tsumoto, K.; Kumagai, I.; Erdmann, V.A. Cell-free expression of two single-chain monoclonal antibodies against lysozyme: Effect of domain arrangement on the expression. J. Biochem. (Tokyo) 1999, 125, 328–333. [Google Scholar] [CrossRef]
- Jiang, X.; Ookubo, Y.; Fujii, I.; Nakano, H.; Yamane, T. Expression of Fab fragment of catalytic antibody 6D9 in an Escherichia coli in vitro coupled transcription/translation system. FEBS Lett. 2002, 514, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Swartz, J.R. Enhancing multiple disulfide bonded protein folding in a cell-free system. Biotechnol. Bioeng. 2004, 86, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.-S.; Kim, D.-M.; Kim, T.-W.; Park, C.-G.; Choi, C.-Y. Providing an oxidizing environment for the cell-free expression of disulfide-containing proteins by exhausting the reducing activity of Escherichia coli S30 extract. Biotechnol. Progr. 2006, 22, 1225–1228. [Google Scholar] [CrossRef]
- Goerke, A.R.; Swartz, J.R. Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol. Bioeng. 2008, 99, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Knapp, K.G.; Goerke, A.R.; Swartz, J.R. Cell-free synthesis of proteins that require disulfide bonds using glucose as an energy source. Biotechnol. Bioeng. 2007, 97, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Groff, D.; Armstrong, S.; Rivers, P.J.; Zhang, J.; Yang, J.; Green, E.; Rozzelle, J.; Liang, S.; Kittle, J.D.; Steiner, A.R.; et al. Engineering toward a bacterial "endoplasmic reticulum" for the rapid expression of immunoglobulin proteins. mAbs 2014, 6, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Bulleid, N.J.; Bassel-Duby, R.S.; Freedman, R.B.; Sambrook, J.F.; Gething, M.J. Cell-free synthesis of enzymically active tissue-type plasminogen activator. Protein folding determines the extent of N-linked glycosylation. Biochem. J. 1992, 286, 275–280. [Google Scholar] [PubMed]
- Ezure, T.; Suzuki, T.; Shikata, M.; Ito, M.; Ando, E.; Nishimura, O.; Tsunasawa, S. Expression of proteins containing disulfide bonds in an insect cell-free system and confirmation of their arrangements by MALDI-TOF MS. Proteomics 2007, 24, 4424–4434. [Google Scholar] [CrossRef]
- Kubick, S.; Schacherl, J.; Fleischer-Notter, H.; Royall, E.; Roberts, L.O.; Stiege, W. In vitro translation in an insect-based cell-free system. In Cell-Free Protein Expression; Swartz, J.R., Ed.; Springer: Berlin, Germany, 2003; pp. 209–217. [Google Scholar]
- Kubick, S.; Gerrits, M.; Merk, H.; Stiege, W.; Erdmann, V.A. In vitro synthesis of posttranslationally modified membrane proteins. In "Membrane Protein Crystallization" Current Topics in Membranes; DeLucas, L., Ed.; Elsevier: Burlington, Canada, 2009; Chapter 2; pp. 25–49. [Google Scholar]
- Sachse, R.; Wüstenhagen, D.; Šamalíková, M.; Gerrits, M.; Bier, F.F.; Kubick, S. Synthesis of membrane proteins in eukaryotic cell-free systems. Eng. Life Sci. 2013, 13, 39–48. [Google Scholar] [CrossRef]
- Dondapati, S.K.; Kreir, M.; Quast, R.B.; Wüstenhagen, D.A.; Brüggemann, A.; Fertig, N.; Kubick, S. Membrane assembly of the functional KcsA potassium channel in a vesicle-based eukaryotic cell-free translation system. Biosens. Bioelectron. 2014, 59, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Quast, R.B.; Sachse, R.; Schulze, C.; Wüstenhagen, D.A.; Kubick, S. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS One 2014, 9, e96635. [Google Scholar] [CrossRef]
- Schulze-Gahmen, U.; Rini, J.M.; Wilson, I.A. Detailed Analysis of the Free and Bound Conformations of an Antibody: X-ray Structures of Fab 17/9 and Three Different Fab-peptide Complexes. J. Mol. Biol. 1993, 234, 1098–1118. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Haun, Y.; Deng, J.; Gao, F.; Pan, B.; Cui, D. Expression of Single-Chain Fv Gene Specific for gamma-Seminoprotein by RTS and Its Biological Activity Identification. Biotechnol. Prog. 2006, 22, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Galeffi, P.; Lombardi, A.; Pietraforte, I.; Novelli, F.; Donato, M.D.; Sperandei, M.; Tornambé, A.; Fraioli, R.; Martayan, A.; Natali, P.G.; et al. Functional expression of a single-chain antibody to ErbB-2 in plants and cell-free systems. J. Transl. Med. 2006, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, H.; Hara, T.; Tanimura, R.; Fukuyama, S.; Cagnon, C.; Kohara, A.; Fujii, I. Site-directed mutagenesis of active site contact residues in a hydrolytic abzyme: Evidence for an essential histidine involved in transition state stabilization. J. Mol. Biol. 1997, 267, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, H.; Karaki, Y.; Kikuchi, M.; Fujii, I. Prodrug activation via catalytic antibodies. Proc. Natl. Acad. Sci. USA 1993, 90, 5337–5340. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Suzuki, H.; Fukuba, T.; Jiang, X.; Nakano, H.; Yamane, T. Improvements in the cell-free production of functional antibodies using cell extract from protease-deficient Escherichia coli mutant. J. Biosci. Bioeng. 2005, 99, 181–186. [Google Scholar] [CrossRef]
- Oh, I.-S.; Lee, J.-C.; Lee, M.-S.; Chung, J.-H.; Kim, D.-M. Cell-free production of functional antibody fragments. Bioproc. Biosyst. Eng. 2010, 33, 127–132. [Google Scholar] [CrossRef]
- Frey, S.; Haslbeck, M.; Hainzl, O.; Buchner, J. Synthesis and characterization of a functional intact IgG in a prokaryotic cell-free expression system. Biol. Chem. 2008, 389, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Stafford, R.L.; Matsumoto, M.L.; Yin, G.; Cai, Q.; Fung, J.J.; Stephenson, H.; Gill, A.; You, M.; Lin, S.-H.; Wang, W.D.; et al. In vitro Fab display: A cell-free system for IgG discovery. Protein Eng. Des. Sel. 2014, 27, 97–109. [Google Scholar] [CrossRef]
- Hillebrecht, J.R.; Chong, S. A comparative study of protein synthesis in in vitro systems: From the prokaryotic reconstituted to the eukaryotic extract-based. BMC Biotechnol. 2008, 8, 1790–1793. [Google Scholar] [CrossRef]
- Langlais, C.; Guilleaume, B.; Wermke, N.; Scheuermann, T.; Ebert, L.; LaBaer, J.; Korn, B. A systematic approach for testing expression of human full-length proteins in cell-free expression systems. BMC Biotechnol. 2007, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tarui, H.; Murata, M.; Tani, I.; Imanishi, S.; Nishikawa, S.; Hara, T. Establishment and characterization of translation/glycosylation in insect cell (Spodoptera frugiperda 21) extract prepared with high pressure treatment. Appl. Microbiol. Biotechnol. 2001, 55, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.H.; Huang, C.-J.; Newton, B.S.; Ritter, G.; Old, L.J.; Batt, C.A. Factors affecting endotoxin removal from recombinant therapeutic proteins by anion exchange chromatography. Protein Exp. Purif. 2009, 64, 76–81. [Google Scholar] [CrossRef]
- Gusdon, J.P.J.; Stavitsky, A.B.; Armentrout, S.A. Synthesis of gamma G antibody and immunoglobulin on polyribosomes in a cell-free system. Proc. Natl. Acad. Sci. USA 1967, 58, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Vassalli, P. Studies on cell-free synthesis of rat immunoglobulins, I. A cell-free system for protein synthesis prepared from lymph-node microsomal vesicles. Proc. Natl. Acad. Sci. USA 1967, 58, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Vassalli, P.; Lisowska-Bernstein, B.; Lamm, M.E.; Benacerraf, B. Studies on cell-free synthesis of rat immunoglobulins, II. Synthesis of immunoglobulin and of antibody to the dinitrophenyl hapten. Proc. Natl. Acad. Sci. USA 1967, 58, 2422–2429. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, P.J.; Johnson, V.G.; Andrew, S.M.; Hoogenboom, H.R.; Raus, J.C.; Youle, R.J. Characterization of single-chain antibody (sFv)-toxin fusion proteins produced in vitro in rabbit reticulocyte lysate. J. Biol. Chem. 1993, 268, 5302–5308. [Google Scholar] [PubMed]
- Hanes, J.; Jermutus, L.; Schaffitze, C.; Plückthun, A. Comparison of Escherichia coli and rabbit reticulocyte ribosome display systems. FEBS Lett. 1999, 450, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Merk, H.; Schenk, J.A.; Stöcklein, W.F.M.; Wüstenhagen, D.A.; Micheel, B.; Duschl, C.; Bier, F.F.; Kubick, S. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J. Biotechnol. 2012, 164, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Schenk, J.A.; Sellrie, F.; Böttger, V.; Menning, A.; Stöcklein, W.F.M.; Micheel, B. Generation and application of a fluorescein-specific single chain antibody. Biochimie 2007, 89, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Micheel, B.; Janstcheff, P.; Böttger, V.; Scharte, G.; Kaiser, G.; Stolley, P.; Karawajew, L. The production and radioimmunoassay application of monoclonal antibodies to fluorescein isothiocyanate (FITC). J. Immunol. Methods 1988, 111, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Merk, H.; Gless, C.; Maertens, B.; Gerrits, M.; Stiege, W. Cell-free synthesis of functional and endotoxin-free antibody Fab fragments by translocation into microsomes. BioTechniques 2012, 53, 153–160. [Google Scholar] [PubMed]
- Tu, T.; Weissman, J. Oxidative protein folding in eukaryotes mechanisms and consequences. J. Cell Biol. 2004, 164, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Kapanidis, A.N.; Weiss, S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J. Chem. Phys. 2002, 117, 10953–10964. [Google Scholar] [CrossRef]
- Elvira, C.; Gallardo, A.; Roman, J.; Cifuentes, A. Covalent polymer-drug conjugates. Molecules 2005, 10, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Kanter, G.; Yang, J.; Voloshin, A.; Levy, S.; Swartz, J.R.; Levy, R. Cell-free production of scFv fusion proteins: An efficient approach for personalized lymphoma vaccines. Blood 2007, 109, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.G.; Ng, P.P.; Levy, S.; Levy, R.; Swartz, J.R. Escherichia coli-based production of a tumor idiotype antibody fragment - tetanus toxin fragment C fusion protein vaccine for B cell lymphoma. Protein Exp. Purif. 2011, 75, 15–20. [Google Scholar] [CrossRef]
- Shimizu, Y.; Kuruma, Y.; Ying, B.-W.; Umekage, S.; Ueda, T. Cell-free translation systems for protein engineering. FEBS J. 2006, 273, 4133–4140. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, M.; Strey, J.; Claußnitzer, I.; von Groll, U.; Schäfer, F.; Rimmele, M.; Stiege, W. Cell-Free synthesis of defined protein conjugates by site-directed cotranslational labeling. In Cell-free Protein Expression; Kudlicki, T., Katzen, F., Bennett, R., Eds.; Landes Bioscience: Austin, TX, USA, 2007. [Google Scholar]
- Budisa, N. Prolegomena to future experimental efforts on genetic code engineering by expanding its amino acid repertoire. Angew. Chem. Int. Ed. 2004, 43, 6426–6463. [Google Scholar] [CrossRef]
- Service, R.F. Unnatural amino acid could prove boon for protein therapeutics. Science 2005, 308, 44. [Google Scholar] [PubMed]
- Mendel, D.; Cornish, V.; Schultz, P. Site-directed mutagenesis with an expanded genetic code. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 435–462. [Google Scholar] [CrossRef] [PubMed]
- Kurzchalia, T.V.; Wiedmann, M.; Breter, H.; Zimmermann, W.; Bauschke, E.; Rapoport, T.A. tRNA-mediated labelling of proteins with biotin. A nonradioactive method for the detection of cell-free translation products. Eur. J. Biochem. 1988, 172, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Quast, R.B.; Claussnitzer, I.; Merk, H.; Kubick, S.; Gerrits, M. Synthesis and site-directed fluorescence labeling of azido proteins using eukaryotic cell-free orthogonal translation systems. Anal. Biochem. 2014, 451, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Schmied, W.H.; Chin, J.W. Reprogramming the genetic code: From triplet to quadruplet codes. Angew. Chem. Int. Ed. 2012, 51, 2288–2297. [Google Scholar] [CrossRef]
- Sisido, M.; Ninomiya, K.; Ohtsuki, T.; Hohsaka, T. Four-base codon/anticodon strategy and non-enzymatic aminoacylation for protein engineering with non-natural amino acids. Methods 2005, 36, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.G.; Ng, P.P.; Kuo, C.-C.; Levy, S.; Levy, R.; Swartz, J.R. Cell-free production of Gaussia princeps luciferase—antibody fragment bioconjugates for ex vivo detection of tumor cells. Biochem. Biophys. Res. Commun. 2009, 390, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Hallam, T.; Smider, V. Unnatural amino acids in novel antibody conjugates. Future Med. Chem. 2014, 6, 1309–1324. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, E.S.; Heibeck, T.H.; Gill, A.; Li, X.; Murray, C.J.; Madlansacay, M.R.; Tran, C.; Uter, N.T.; Yin, G.; Rivers, P.J.; et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug. Chem. 2014, 25, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Furumoto, S.; Higuchi, K.; Yokoyama, J.; Zhang, M.-R.; Yanai, K.; Iwata, R.; Kigawa, T. Rapid biochemical synthesis of 11C-labeled single chain variable fragment antibody for immuno-PET by cell-free protein synthesis. Bioorg. Med. Chem. 2012, 20, 6579–6582. [Google Scholar] [CrossRef] [PubMed]
- Brödel, A.K.; Sonnabend, A.; Kubick, S. Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 2013, 111, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Brödel, A.K.; Sonnabend, A.; Roberts, L.O.; Stech, M.; Wüstenhagen, D.A.; Kubick, S. IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems. PLoS One 2013, 8, e82234. [Google Scholar] [CrossRef]
- Ghaderi, D.; Zhang, M.; Hurtado-Ziola, N.; Varki, A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol. Genet. Eng. Rev. 2012, 28, 147–175. [Google Scholar] [PubMed]
- Shade, K.-T.C.; Anthony, R.M. Antibody Glycosylation and Inflammation. Antibodies 2013, 2, 92–414. [Google Scholar] [CrossRef]
- Jefferis, R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009, 8, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Guarino, C.; DeLisa, M. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology 2012, 22, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Kigawa, T.; Yabuki, T.; Yoshida, Y.; Tsutsui, M.; Ito, Y.; Shibata, T.; Yokoyama, S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999, 442, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Madin, K.; Sawasaki, T.; Ogasawara, T.; Endo, Y. A highly efficient and robust protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Ramya, R.; Subramanian, B.M.; Sivakumar, V.; Senthilkumar, R.L.; Rao, K.R.S.S.; Srinivasan, V.A. Expression and solubilization of insect cell-based rabies virus glycoprotein and assessment of its immunogenicity and protective efficacy in mice. Clin. Vaccine Immunol. 2011, 18, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Naftzger, C.; Takechi, Y.; Kohda, H.; Hara, I.; Vijayasaradhi, S.; Houghton, A.N. Immune response to a differentiation antigen induced by altered antigen: A study of tumor rejection and autoimmunity. Proc. Natl. Acad. Sci. USA 1996, 93, 14809–14814. [Google Scholar] [CrossRef] [PubMed]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Lusic, H.; Neumann, H.; Kapadnis, P.B.; Deiters, A.; Chin, J.W. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA synthetase/tRNACUA pair and click chemistry. J. Am. Chem. Soc. 2009, 131, 8720–8721. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, J.B.B.; Presta, L.G.; Carter, P. "Knobs-into-holes" engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996, 9, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lee, J.; Tran, C.; Heibeck, T.H.; Wang, W.D.; Yang, J.; Stafford, R.L.; Steiner, A.R.; Sato, A.K.; Hallam, T.J.; et al. Production of bispecific antibodies in "Knobs-into-Holes" using a cell-free expression system. mAbs 2014. [Google Scholar] [CrossRef]
- Murray, C.J.; Baliga, R. Cell-free translation of peptides and proteins: From high throughput screening to clinical production. Curr. Opin. Chem. Biol. 2013, 17, 420–426. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stech, M.; Kubick, S. Cell-Free Synthesis Meets Antibody Production: A Review. Antibodies 2015, 4, 12-33. https://doi.org/10.3390/antib4010012
Stech M, Kubick S. Cell-Free Synthesis Meets Antibody Production: A Review. Antibodies. 2015; 4(1):12-33. https://doi.org/10.3390/antib4010012
Chicago/Turabian StyleStech, Marlitt, and Stefan Kubick. 2015. "Cell-Free Synthesis Meets Antibody Production: A Review" Antibodies 4, no. 1: 12-33. https://doi.org/10.3390/antib4010012




