Efforts and Challenges in Engineering the Genetic Code
Abstract
:1. Introduction
2. Genetic Code Engineering
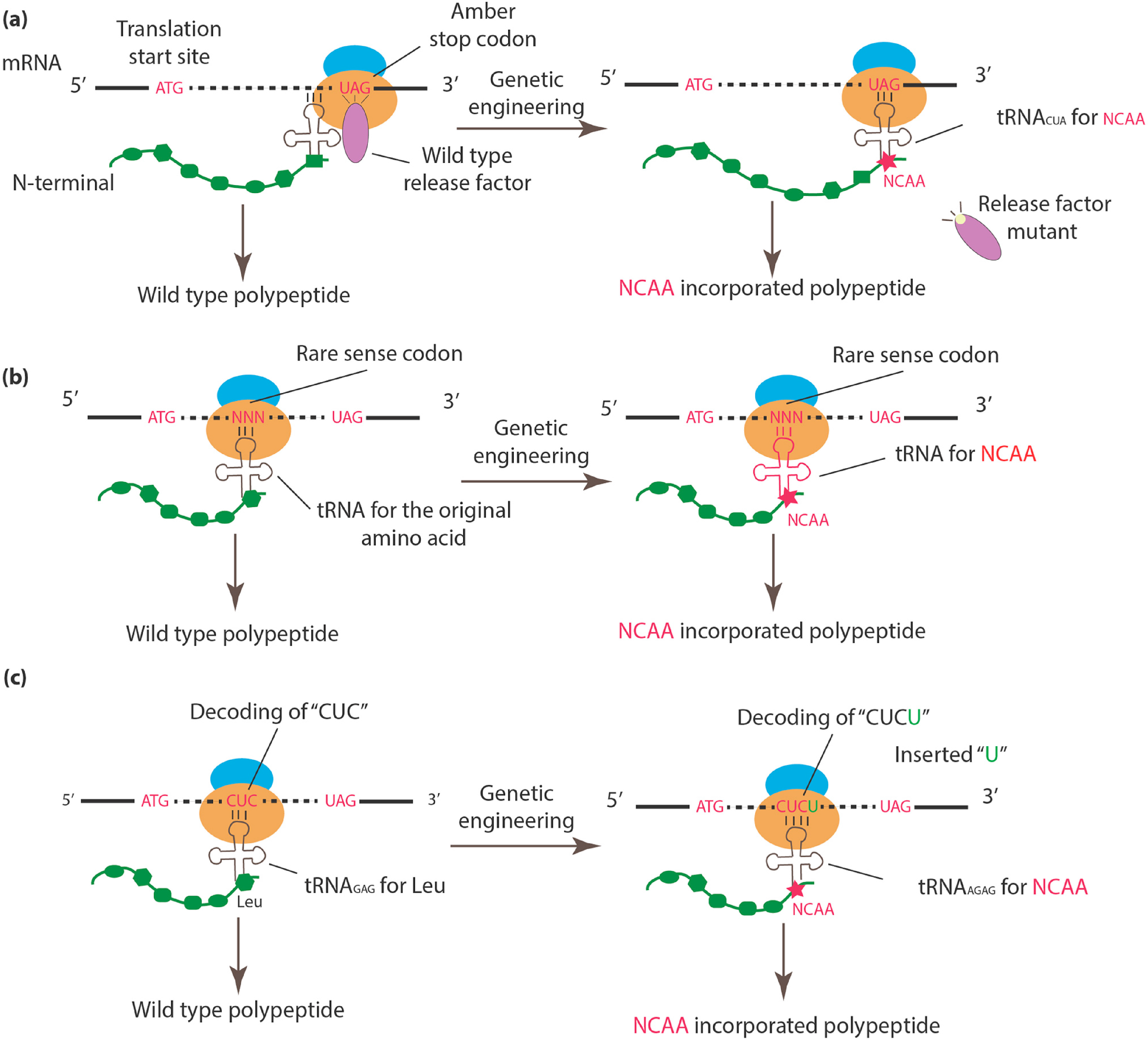
2.1. Incorporation of NCAAs into Specific Sites
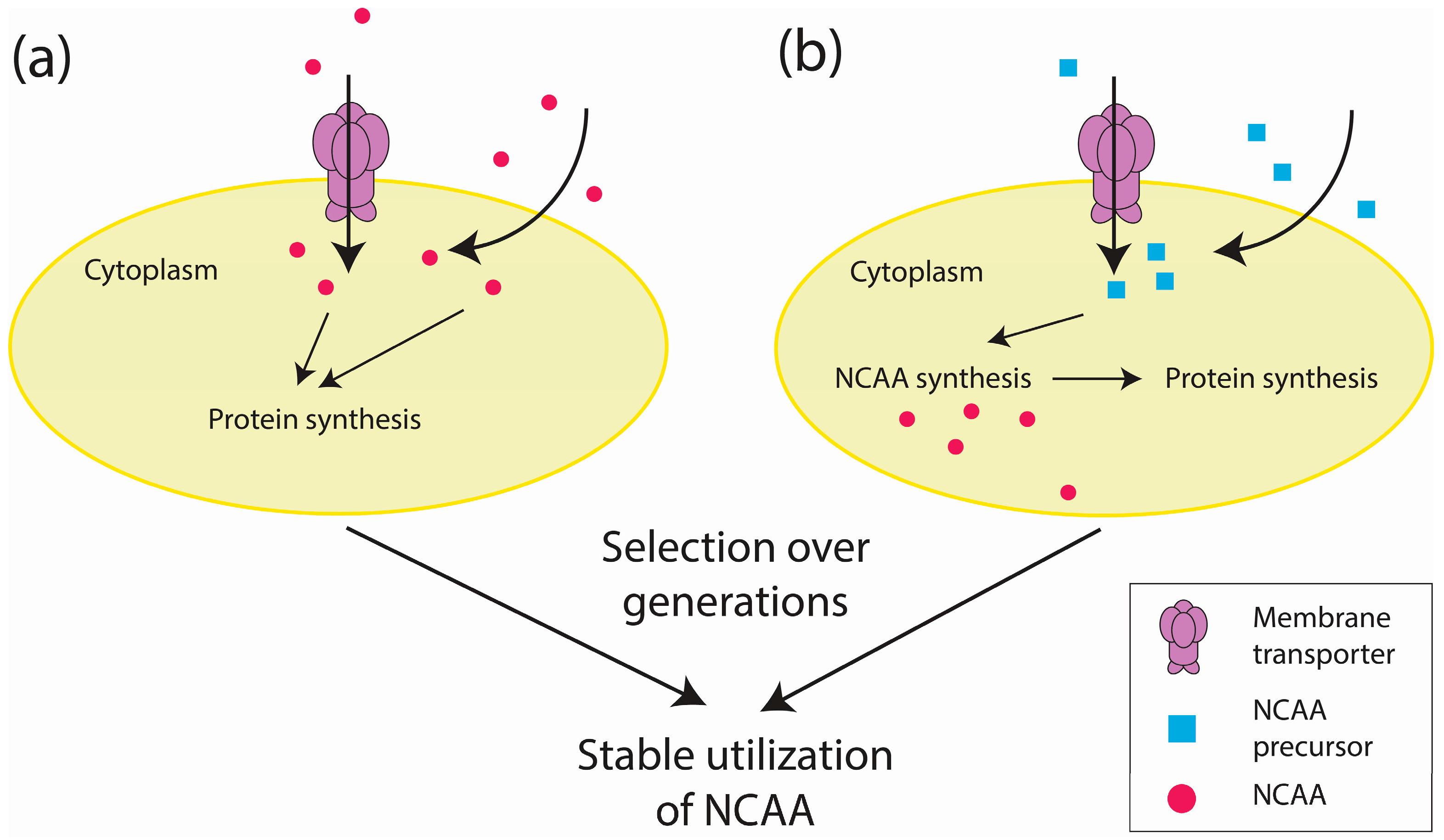
2.2. Proteome-Wide Incorporation of NCAAs
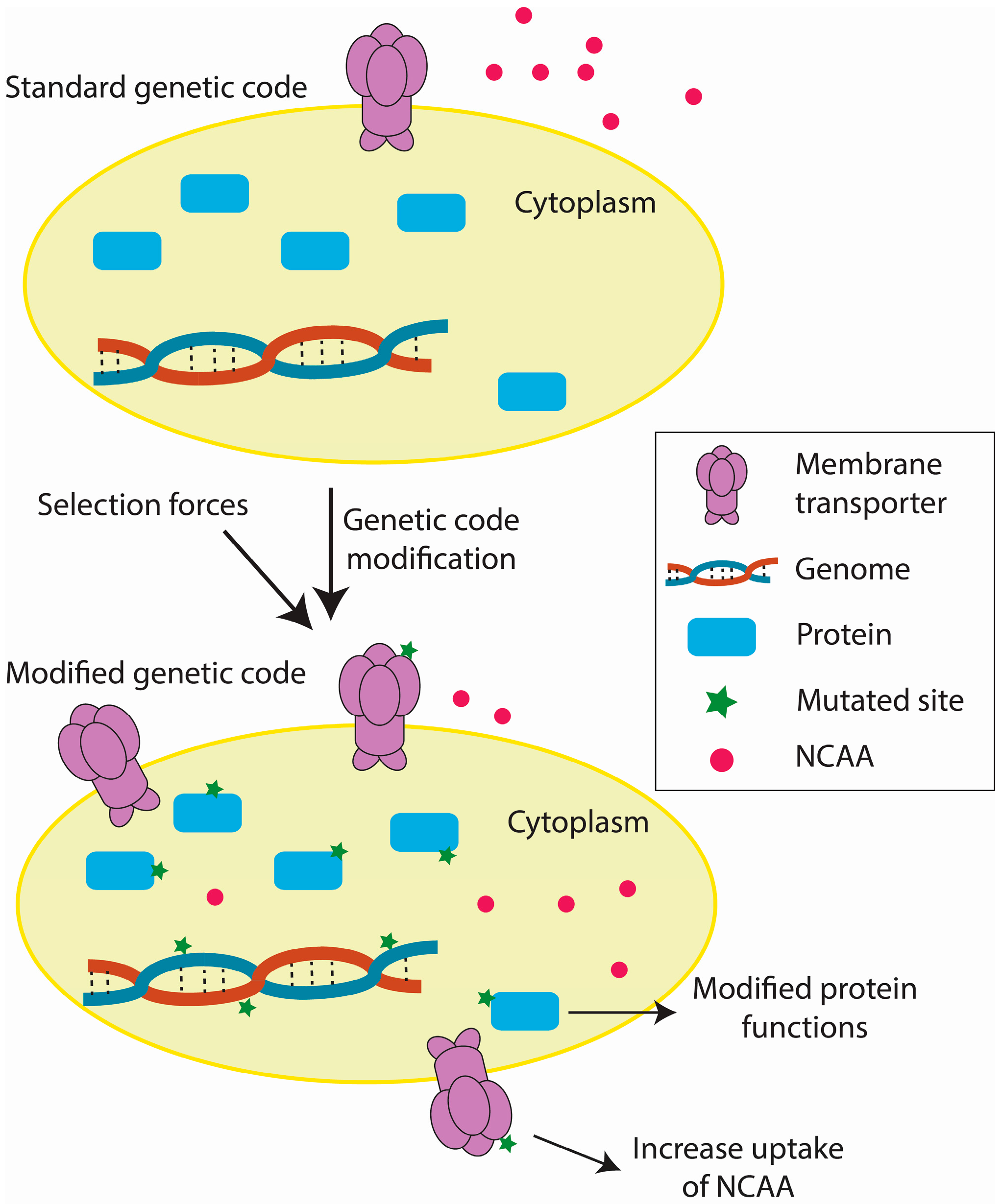
3. Challenges of Genetic Code Engineering
3.1. Inhibitory Effects of Engineered Genetic Codes
3.2. Discovering the Key Genes Controlling the Genetic Code
3.3. Lack of Transcriptomic and Proteomic Studies Related to Engineered Genetic Codes
3.4. Environmental Factors Affecting Adaptation to Engineered Genetic Codes
4. Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Cone, J.E.; Del Río, R.M.; Davis, J.N.; Stadtman, T.C. Chemical characterization of the selenoprotein component of clostridial glycine reductase: Identification of selenocysteine as the organoselenium moiety. Proc. Natl. Acad. Sci. USA 1976, 73, 2659–2663. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Gong, W.; Ferguson, T.K.; James, C.M.; Krzycki, J.A.; Chan, M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 2002, 296, 1462–1466. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, G.; James, C.M.; Krzycki, J.A. Pyrrolysine encoded by UAG in archaea: Charging of a UAG-decoding specialized tRNA. Science 2002, 296, 1459–1462. [Google Scholar] [CrossRef] [PubMed]
- Adachi, M.; Cavalcanti, A.R.O. Tandem stop codons in ciliates that reassign stop codons. J. Mol. Evol. 2009, 68, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Beznosková, P.; Gunišová, S.; Valášek, L.S. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA 2016, 22, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Osawa, S.; Jukes, T.H. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 1989, 28, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.W.; Yarus, M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994, 235, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Andersson, S.G.; Kurland, C.G. Genomic evolution drives the evolution of the translation system. Biochem. Cell Biol. 1995, 73, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Söll, D.; RajBhandary, U.L. The genetic code—Thawing the “frozen accident”. J. Biosci. 2006, 31, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Novozhilov, A.S. Origin and evolution of the genetic code: The universal enigma. IUBMB Life 2009, 61, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Hartman, M.C.T.; Josephson, K.; Lin, C.-W.; Szostak, J.W. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE 2007, 2, e972. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.; Hennecke, H. Relaxing the substrate specificity of an aminoacyl-tRNA synthetase allows in vitro and in vivo synthesis of proteins containing unnatural amino acids. FEBS Lett. 1995, 364, 272–275. [Google Scholar] [CrossRef]
- Xu, Z.J.; Love, M.L.; Ma, L.Y.; Blum, M.; Bronskill, P.M.; Bernstein, J.; Grey, A.A.; Hofmann, T.; Camerman, N.; Wong, J.T. Tryptophanyl-tRNA synthetase from Bacillus subtilis. Characterization and role of hydrophobicity in substrate recognition. J. Biol. Chem. 1989, 264, 4304–4311. [Google Scholar] [PubMed]
- Lovett, P.S.; Ambulos, N.P.; Mulbry, W.; Noguchi, N.; Rogers, E.J.; Rogers, E.J. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J. Bacteriol. 1991, 173, 1810–1812. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, F.; Iglesias-Groth, S.; Angelini, G.; Hafez, Y. Stability toward high energy radiation of non-proteinogenic amino acids: Implications for the origins of life. Life 2013, 3, 449–473. [Google Scholar] [CrossRef] [PubMed]
- Richmond, M.H. The effect of amino acid analogues on growth and protein synthesis in microorganisms. Bacteriol. Rev. 1962, 26, 398–420. [Google Scholar] [PubMed]
- Lea, P.J.; Norris, R.D. The use of amino acid analogues in studies on plant metabolism. Phytochemistry 1976, 15, 585–595. [Google Scholar] [CrossRef]
- Rodgers, K.J.; Hume, P.M.; Dunlop, R.A.; Dean, R.T. Biosynthesis and turnover of DOPA-containing proteins by human cells. Free Radic. Biol. Med. 2004, 37, 1756–1764. [Google Scholar] [CrossRef] [PubMed]
- Commans, S.; Böck, A. Selenocysteine inserting tRNAs: An overview. FEMS Microbiol. Rev. 1999, 23, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.J.; Banu, L.; Chen, Y.; Mandel, S.J.; Kieffer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a selenocysteine codon in Type I deiodinase requires sequences in the 3′ untranslated region. Nature 1991, 353, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.M.; Paredes, J.A.; Moura, G.R.; Manadas, B.; Lima-Costa, T.; Rocha, R.; Miranda, I.; Gomes, A.C.; Koerkamp, M.J.G.; Perrot, M.; et al. Critical roles for a genetic code alteration in the evolution of the genus Candida. EMBO J. 2007, 26, 4555–4565. [Google Scholar] [CrossRef] [PubMed]
- Krzycki, J.A. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 2004, 8, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Polycarpo, C.; Ambrogelly, A.; Bérubé, A.; Winbush, S.M.; McCloskey, J.A.; Crain, P.F.; Wood, J.L.; Söll, D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA 2004, 101, 12450–12454. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Rother, M.; Krzycki, J.A. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea 2010, 2010, 453642. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; Hughes, R.A.; Tze-Fei Wong, J.; Ellington, A.D. Evolving new genetic codes. Trends Ecol. Evol. 2004, 19, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hammerling, M.J.; Ellefson, J.W.; Boutz, D.R.; Marcotte, E.M.; Ellington, A.D.; Barrick, J.E. Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 2014, 10, 178–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Hohn, M.J.; Umehara, T.; Guo, L.-T.; Osborne, E.M.; Benner, J.; Noren, C.J.; Rinehart, J.; Söll, D. Expanding the genetic code of Escherichia coli with phosphoserine. Science 2011, 333, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yanagisawa, T.; Ohtake, K.; Wakamori, M.; Adachi, J.; Hino, N.; Sato, A.; Kobayashi, T.; Hayashi, A.; Shirouzu, M.; et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem. Biophys. Res. Commun. 2011, 411, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Bacher, J.M.; Ellington, A.D. Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 2001, 183, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.T. Membership mutation of the genetic code: Loss of fitness by tryptophan. Proc. Natl. Acad. Sci. USA 1983, 80, 6303–6306. [Google Scholar] [CrossRef] [PubMed]
- Mat, W.K.; Xue, H.; Wong, J.T.F. Genetic code mutations: The breaking of a three billion year invariance. PLoS ONE 2010, 5, e12206. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.R.; Li, Z.; Lewis, A.O.; Isaacs, F.J. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat. Protoc. 2014, 9, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Ostrov, N.; Landon, M.; Guell, M.; Kuznetsov, G.; Teramoto, J.; Cervantes, N.; Zhou, M.; Singh, K.; Napolitano, M.G.; Moosburner, M.; et al. Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Budisa, N.; Völler, J.-S.; Koksch, B.; Acevedo-Rocha, C.G.; Kubyshkin, V.; Agostini, F. Xenobiology meets enzymology: Exploring the potential of unnatural building blocks in biocatalysis. Angew. Chem. Int. Ed. 2017. [Google Scholar] [CrossRef]
- Wang, L.; Brock, A.; Schultz, P.G. Adding l-3-(2-Naphthyl)alanine to the genetic code of E. coli. J. Am. Chem. Soc. 2002, 124, 1836–1837. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W.; Martin, A.B.; King, D.S.; Wang, L.; Schultz, P.G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 11020–11024. [Google Scholar] [CrossRef] [PubMed]
- Scolnick, E.; Tompkins, R.; Caskey, T.; Nirenberg, M. Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. USA 1968, 61, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Petry, S.; Brodersen, D.E.; Murphy, F.V.; Dunham, C.M.; Selmer, M.; Tarry, M.J.; Kelley, A.C.; Ramakrishnan, V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 2005, 123, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.F.; Xu, J.; Shen, Z.; Takimoto, J.K.; Schultz, M.D.; Schmitz, R.J.; Xiang, Z.; Ecker, J.R.; Briggs, S.P.; Wang, L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011, 7, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.; Lercher, L.; Spicer, C.D.; Davis, B.G. Designing logical codon reassignment—Expanding the chemistry in biology. Chem. Sci. 2015, 6, 50–69. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, W.; Liu, W.R. Towards reassigning the rare AGG codon in Escherichia coli. ChemBioChem 2014, 15, 1750–1754. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yamaguchi, A.; Ohtake, K.; Takahashi, M.; Hayashi, A.; Iraha, F.; Kira, S.; Yanagisawa, T.; Yokoyama, S.; Hoshi, H.; et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015, 43, 8111–8122. [Google Scholar] [CrossRef] [PubMed]
- Bohlke, N.; Budisa, N. Sense codon emancipation for proteome-wide incorporation of noncanonical amino acids: Rare isoleucine codon AUA as a target for genetic code expansion. FEMS Microbiol. Lett. 2014, 351, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.C.; Wu, N.; Santoro, S.W.; Lakshman, V.; King, D.S.; Schultz, P.G. An expanded genetic code with a functional quadruplet codon. Proc. Natl. Acad. Sci. USA 2004, 101, 7566–7571. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.A.; Lester, H.A.; Dougherty, D.A. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc. Natl. Acad. Sci. USA 2006, 103, 8650–8655. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Schultz, P.G.; Guo, J. An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem. Biol. 2013, 8, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Magliery, T.J.; Anderson, J.C.; Schultz, P.G. Expanding the genetic code: Selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J. Mol. Biol. 2001, 307, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.C.; Magliery, T.J.; Schultz, P.G. Exploring the limits of codon and anticodon size. Chem. Biol. 2002, 9, 237–244. [Google Scholar] [CrossRef]
- Taki, M.; Matsushita, J.; Sisido, M. Expanding the genetic code in a mammalian cell line by the introduction of four-base codon/anticodon pairs. ChemBioChem 2006, 7, 425–428. [Google Scholar] [CrossRef] [PubMed]
- Seligmann, H. Natural chymotrypsin-like-cleaved human mitochondrial peptides confirm tetra-, pentacodon, non-canonical RNA translations. Biosystems 2016, 147, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007, 25, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Kast, P.; Hennecke, H. Amino acid substrate specificity of Escherichia coli phenylalanyl-tRNA synthetase altered by distinct mutations. J. Mol. Biol. 1991, 222, 99–124. [Google Scholar] [CrossRef]
- Hoesl, M.G.; Oehm, S.; Durkin, P.; Darmon, E.; Peil, L.; Aerni, H.-R.; Rappsilber, J.; Rinehart, J.; Leach, D.; Söll, D.; et al. Chemical evolution of a bacterial proteome. Angew. Chem. Int. Ed. 2015, 54, 10030–10034. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, M.; Gattner, M.J.; Viverge, B.; Bretzler, J.; Eisen, D.; Stadlmeier, M.; Vrabel, M.; Carell, T. Orchestrating the biosynthesis of an unnatural pyrrolysine amino acid for its direct incorporation into proteins inside living cells. Chem. Eur. J. 2015, 21, 7701–7704. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Biava, H.; Contestabile, R.; Budisa, N.; di Salvo, M. Coupling bioorthogonal chemistries with artificial metabolism: Intracellular biosynthesis of azidohomoalanine and its incorporation into recombinant proteins. Molecules 2014, 19, 1004–1022. [Google Scholar] [CrossRef] [PubMed]
- Exner, M.P.; Kuenzl, T.; To, T.M.T.; Ouyang, Z.; Schwagerus, S.; Hoesl, M.G.; Hackenberger, C.P.R.; Lensen, M.C.; Panke, S.; Budisa, N. Design of S-allylcysteine in situ production and incorporation based on a novel pyrrolysyl-tRNA synthetase variant. Chembiochem 2017, 18, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.; Uno, T.; Chiu, H.-P.; Grunewald, J.; Cellitti, S.E.; Crossgrove, T.; Hao, X.; Fan, Q.; Quinn, L.L.; Patterson, P.; et al. Site-specific protein modifications through pyrroline-carboxy-lysine residues. Proc. Natl. Acad. Sci. USA 2011, 108, 10437–10442. [Google Scholar] [CrossRef] [PubMed]
- Mehl, R.A.; Anderson, J.C.; Santoro, S.W.; Wang, L.; Martin, A.B.; King, D.S.; Horn, D.M.; Schultz, P.G. Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc. 2003, 125, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Rocha, C.G.; Budisa, N. Xenomicrobiology: A roadmap for genetic code engineering. Microb. Biotechnol. 2016, 9, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Völler, J.-S.; Budisa, N. Coupling genetic code expansion and metabolic engineering for synthetic cells. Curr. Opin. Biotechnol. 2017, 48, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.M.; Brown, A.S.; Corwin, L.M.; Troxler, R.F.; Franzblau, C. Effect of l-azetidine 2-carboxylic acid on growth and proline metabolism in Escherichia coli. Biochim. Biophys. Acta 1975, 404, 180–187. [Google Scholar] [CrossRef]
- Unger, L.; DeMoss, R.D. Action of a proline analogue, l-thiazolidine-4-carboxylic acid, in Escherichia coli. J. Bacteriol. 1966, 91, 1556–1563. [Google Scholar] [PubMed]
- Moran, S.; Rai, D.K.; Clark, B.R.; Murphy, C.D. Precursor-directed biosynthesis of fluorinated iturin A in Bacillus spp. Org. Biomol. Chem. 2009, 7, 644. [Google Scholar] [CrossRef] [PubMed]
- Téllez, R.; Jacob, G.; Basilio, C.; George-Nascimento, C. Effect of ethionine on the in vitro synthesis and degradation of mitochondrial translation products in yeast. FEBS Lett. 1985, 192, 88–94. [Google Scholar] [CrossRef]
- Rosenthal, G.; Lambert, J.; Hoffmann, D. Canavanine incorporation into the antibacterial proteins of the fly, Phormia terranovae (Diptera), and its effect on biological activity. J. Biol. Chem. 1989, 26417, 9768–9771. [Google Scholar]
- Teramoto, H.; Kojima, K. Incorporation of methionine analogues into bombyx mori silk fibroin for click modifications. Macromol. Biosci. 2015, 15, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Poirson-Bichat, F.; Lopez, R.; Bras Gonçalves, R.A.; Miccoli, L.; Bourgeois, Y.; Demerseman, P.; Poisson, M.; Dutrillaux, B.; Poupon, M.F. Methionine deprivation and methionine analogs inhibit cell proliferation and growth of human xenografted gliomas. Life Sci. 1997, 60, 919–931. [Google Scholar] [CrossRef]
- Merkel, L.; Budisa, N. Organic fluorine as a polypeptide building element: In vivo expression of fluorinated peptides, proteins and proteomes. Org. Biomol. Chem. 2012, 10, 7241. [Google Scholar] [CrossRef] [PubMed]
- Schlesinger, S. The effect of amino acid analogues on alkaline phosphatase. Formation in Escherichia coli K-II. Replacement of tryptophan by azatryptophan and by tryptazan. J. Biol. Chem. 1968, 243, 3877–3883. [Google Scholar] [PubMed]
- Pine, M.J. Comparative physiological effects of incorporated amino acid analogs in Escherichia coli. Antimicrob. Agents Chemother. 1978, 13, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.; Kwon, I. Effects of non-natural amino acid incorporation into the enzyme core region on enzyme structure and function. Int. J. Mol. Sci. 2015, 16, 22735–22753. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.; Tirrell, D.A. Site-specific incorporation of tryptophan analogues into recombinant proteins in bacterial cells. 2007, 129, 10431–10437. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.C.-S.; Yim, A.K.-Y.; Mat, W.-K.; Tong, A.H.-Y.; Lok, S.; Xue, H.; Tsui, S.K.-W.; Wong, J.T.-F.; Chan, T.-F. Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol. Evol. 2014, 6, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Gollnick, P.; Ishino, S.; Kuroda, M.I.; Henner, D.J.; Yanofsky, C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1990, 87, 8726–8730. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 2007, 13, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Sarsero, J.P.; Merino, E.; Yanofsky, C. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 2000, 182, 2329–2331. [Google Scholar] [CrossRef] [PubMed]
- Yakhnin, H.; Yakhnin, A.V.; Babitzke, P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol. 2006, 61, 1252–1266. [Google Scholar] [CrossRef] [PubMed]
- Yakhnin, H.; Zhang, H.; Yakhnin, A.V.; Babitzke, P. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 2004, 186, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Tarpey, R.; Babitzke, P. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 1997, 179, 2582–2586. [Google Scholar] [CrossRef] [PubMed]
- Honoré, N.; Cole, S.T. Nucleotide sequence of the aroP gene encoding the general aromatic amino acid transport protein of Escherichia coli K-12: Homology with yeast transport proteins. Nucleic Acids Res. 1990, 18, 653. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.G.; Fan, C.S.; Wu, Y.Q.; Jin, R.L.; Liu, D.X.; Shang, L.; Jiang, P.H. Regulation of aroP expression by tyrR gene in Escherichia coli. Acta Biochim. Biophys. Sin. 2003, 35, 993–997. [Google Scholar] [PubMed]
- Hammerling, M.J.; Gollihar, J.; Mortensen, C.; Alnahhas, R.N.; Ellington, A.D.; Barrick, J.E. Expanded genetic codes create new mutational routes to rifampicin resistance in Escherichia coli. Mol. Biol. Evol. 2016, 33, 2054–2063. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Borrel, G.; Gaci, N.; Peyret, P.; O'Toole, P.W.; Gribaldo, S.; Brugère, J.F. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: Implications for the evolution of a genetic code expansion cassette. Archaea 2014, 2014, 374146. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, P.; Prat, L.; Kucklick, M.; Schäfer, J.G.; Riedel, K.; Rinehart, J.; Söll, D.; Heinemann, I.U. Reducing the genetic code induces massive rearrangement of the proteome. Proc. Natl. Acad. Sci. USA 2014, 111, 17206–17211. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.S.; Ostapchuk, P.; Hearing, P.; Carrico, I.S. Unnatural amino acid incorporation onto adenoviral (Ad) coat proteins facilitates chemoselective modification and retargeting of Ad type 5 vectors. J. Virol. 2011, 85, 7546–7554. [Google Scholar] [CrossRef] [PubMed]
- Tack, D.S.; Ellefson, J.W.; Thyer, R.; Wang, B.; Gollihar, J.; Forster, M.T.; Ellington, A.D. Addicting diverse bacteria to a noncanonical amino acid. Nat. Chem. Biol. 2016, 12, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Prat, L.; Heinemann, I.U.; Aerni, H.R.; Rinehart, J.; O’Donoghue, P.; Söll, D. Carbon source-dependent expansion of the genetic code in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 21070–21075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.H.; Otting, G.; Jackson, C.J. Protein engineering with unnatural amino acids. Curr. Opin. Struct. Biol. 2013, 23, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M. Xenobiology: A new form of life as the ultimate biosafety tool. Bioessays 2010, 32, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sismour, A.M.; Sheng, P.; Puskar, N.L.; Benner, S.A. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007, 35, 4238–4249. [Google Scholar] [CrossRef] [PubMed]
- Sismour, A.M.; Lutz, S.; Park, J.-H.; Lutz, M.J.; Boyer, P.L.; Hughes, S.H.; Benner, S.A. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 2004, 32, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hutter, D.; Sheng, P.; Sismour, A.M.; Benner, S.A. Artificially expanded genetic information system: A new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006, 34, 6095–6101. [Google Scholar] [CrossRef] [PubMed]
- Leconte, A.M.; Hwang, G.T.; Matsuda, S.; Capek, P.; Hari, Y.; Romesberg, F.E. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J. Am. Chem. Soc. 2008, 130, 2336–2343. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, C.A.; Chuang, R.-Y.; Noskov, V.N.; Assad-Garcia, N.; Deerinck, T.J.; Ellisman, M.H.; Gill, J.; Kannan, K.; Karas, B.J.; Ma, L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Yu, A.C.S.; Chan, T.F. Efforts and Challenges in Engineering the Genetic Code. Life 2017, 7, 12. https://doi.org/10.3390/life7010012
Lin X, Yu ACS, Chan TF. Efforts and Challenges in Engineering the Genetic Code. Life. 2017; 7(1):12. https://doi.org/10.3390/life7010012
Chicago/Turabian StyleLin, Xiao, Allen Chi Shing Yu, and Ting Fung Chan. 2017. "Efforts and Challenges in Engineering the Genetic Code" Life 7, no. 1: 12. https://doi.org/10.3390/life7010012
APA StyleLin, X., Yu, A. C. S., & Chan, T. F. (2017). Efforts and Challenges in Engineering the Genetic Code. Life, 7(1), 12. https://doi.org/10.3390/life7010012





