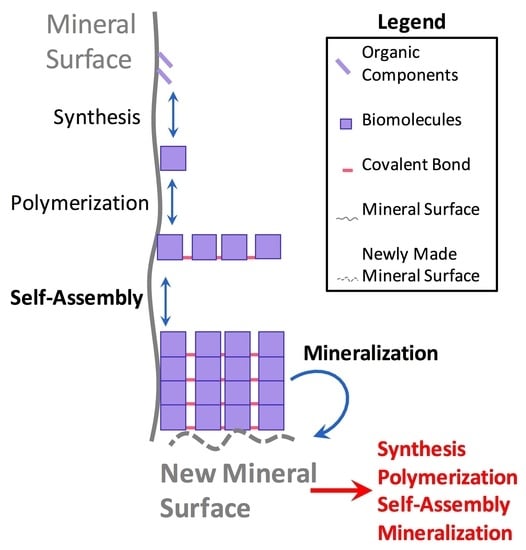
Mineral Surface-Templated Self-Assembling Systems: Case Studies from Nanoscience and Surface Science towards Origins of Life Research
Abstract
:1. Introduction
2. Case Studies
2.1. Simple Organic Molecules
2.2. Nucleic Acids
2.3. Peptides
2.4. Biomineralization
3. Prospective
3.1. Synergistic Cyclical Model of Mineral-Templated Self-Assembling Systems Promoting Mineralization
3.2. Incorporating Recent Discoveries in Nanoscience and Surface Science into Origins of Life Research
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Damineli, A.; Damineli, D.S.C. Origens da vida. Estudos Avançados 2007, 21, 263–284. [Google Scholar] [CrossRef]
- Kawai, S.; Haapasilta, V.; Lindner, B.D.; Tahara, K.; Spijker, P.; Buitendijk, J.A.; Pawlak, R.; Meier, T.; Tobe, Y.; Foster, A.S.; et al. Thermal control of sequential on-surface transformation of a hydrocarbon molecule on a copper surface. Nat. Commun. 2016, 7, 12711. [Google Scholar] [CrossRef] [PubMed]
- Johlin, J.M. Interfacial adsorption as a Function of the Concentration of Colloidal Solutions. J. Biol. Chem. 1929, 84, 543–551. [Google Scholar]
- Nič, M.; Jirát, J.; Košata, B.; Jenkins, A.; McNaught, A. Adsorption. In IUPAC Compendium of Chemical Terminology; Nič, M., Jirát, J., Košata, B., Jenkins, A., McNaught, A., Eds.; IUPAC: Zurich, Switzerland, 2009; ISBN 9780967855097. [Google Scholar]
- Pang, S.H.; Medlin, J.W. Controlling Catalytic Selectivity via Adsorbate Orientation on the Surface: From Furfural Deoxygenation to Reactions of Epoxides. J. Phys. Chem. Lett. 2015, 6, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Akash, R.; Kamalnath, K.; Ahmad, R.; Singh, A.K.; Ravishankar, N. Orientation Selection during Heterogeneous Nucleation: Implications for Heterogeneous Catalysis. J. Phys. Chem. C 2017, 121, 10027–10037. [Google Scholar] [CrossRef]
- Nørskov, J.K. Electronic factors in catalysis. Prog. Surf. Sci. 1991, 38, 103–144. [Google Scholar] [CrossRef]
- Xu, Y.; Schoonen, M.A.A. The absolute energy positions of conduction and valence bands of selected semiconducting minerals. Am. Mineral. 2000, 85, 543–556. [Google Scholar] [CrossRef]
- Hazen, R.M.; Grew, E.S.; Downs, R.T.; Golden, J.; Hystad, G. Mineral Ecology: Chance and Necessity in the Mineral Diversity of Terrestrial Planets. Can. Mineral. 2015, 53, 295–324. [Google Scholar] [CrossRef]
- Schwartz, A.W.; Veen, M.; Bisseling, T.; Chittenden, G.J.F. Prebiotic nucleotide synthesis-demonstration of a geologically plausible pathway. Orig. Life 1975, 6, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Pasek, M.A. Schreibersite on the early Earth: Scenarios for prebiotic phosphorylation. Geosci. Front. 2017, 8, 329–335. [Google Scholar] [CrossRef]
- Gull, M.; Mojica, M.A.; Fernández, F.M.; Gaul, D.A.; Orlando, T.M.; Liotta, C.L.; Pasek, M.A. Nucleoside phosphorylation by the mineral schreibersite. Sci. Rep. 2015, 5, 17198. [Google Scholar] [CrossRef] [PubMed]
- Saladino, R.; Barontini, M.; Cossetti, C.; Di Mauro, E.; Crestini, C. The effects of borate minerals on the synthesis of nucleic acid bases, amino acids and biogenic carboxylic acids from formamide. Orig. Life Evol. Biosph. 2011, 41, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Ricardo, A.; Carrigan, M.A.; Olcott, A.N.; Benner, S.A. Borate minerals stabilize ribose. Science 2004, 303, 196. [Google Scholar] [CrossRef] [PubMed]
- Grew, E.S.; Bada, J.L.; Hazen, R.M. Borate minerals and origin of the RNA world. Orig. Life Evol. Biosph. 2011, 41, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-F. Adsorption and polymerization of amino acids on mineral surfaces: A review. Orig. Life Evol. Biosph. 2008, 38, 211–242. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, H. Adsorption of Nucleic Acid Bases, Ribose, and Phosphate by Some Clay Minerals. Life 2015, 5, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Hedges, J.I.; Hare, P.E. Amino acid adsorption by clay minerals in distilled water. Geochim. Cosmochim. Acta 1987, 51, 255–259. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Philos. Trans. Royal Soc. Lond. B Biol. Sci. 2006, 361, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.P.; Hill, A.R., Jr.; Liu, R.; Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 1996, 381, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Ferris, J.; Ertem, G. Oligomerization of ribonucleotides on montmorillonite: Reaction of the 5′-phosphorimidazolide of adenosine. Science 1992, 257, 1387–1389. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Ferris, J.P. Kinetic and Mechanistic Analysis of Dinucleotide and Oligonucleotide Formation from the 5′-Phosphorimidazolide of Adenosine on Na-Montmorillonite. J. Am. Chem. Soc. 1994, 116, 7564–7572. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. Polymerization on the rocks: Theoretical introduction. Orig. Life Evol. Biosph. 1998, 28, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.R., Jr.; Böhler, C.; Orgel, L.E. Polymerization on the rocks: Negatively-charged alpha-amino acids. Orig. Life Evol. Biosph. 1998, 28, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Orgel, L.E. Polymerization on the rocks: Beta-amino acids and arginine. Orig. Life Evol. Biosph. 1998, 28, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Lahav, N.; White, D.; Chang, S. Peptide formation in the prebiotic era: Thermal condensation of glycine in fluctuating clay environments. Science 1978, 201, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Kitadai, N.; Oonishi, H.; Umemoto, K.; Usui, T.; Fukushi, K.; Nakashima, S. Glycine Polymerization on Oxide Minerals. Orig. Life Evol. Biosph. 2017, 47, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Fujikawa, S.M.; Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 2003, 302, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Hanczyc, M.M.; Mansy, S.S.; Szostak, J.W. Mineral surface directed membrane assembly. Orig. Life Evol. Biosph. 2007, 37, 67–82. [Google Scholar] [CrossRef] [PubMed]
- Sahai, N.; Kaddour, H.; Dalai, P.; Wang, Z.; Bass, G.; Gao, M. Mineral Surface Chemistry and Nanoparticle-aggregation Control Membrane Self-Assembly. Sci. Rep. 2017, 7, 43418. [Google Scholar] [CrossRef] [PubMed]
- Ebisuzaki, T.; Maruyama, S. Nuclear geyser model of the origin of life: Driving force to promote the synthesis of building blocks of life. Geosci. Front. 2017, 8, 275–298. [Google Scholar] [CrossRef]
- Westall, F.; Brack, A. The Importance of Water for Life. Space Sci. Rev. 2018, 214, 50. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell; Garland Science: Oxford, UK, 2014; ISBN 9780815344322. [Google Scholar]
- Grover, M.; He, C.; Hsieh, M.-C.; Yu, S.-S. A Chemical Engineering Perspective on the Origins of Life. Processes 2015, 3, 309–338. [Google Scholar] [CrossRef]
- Mandal, D.; Nasrolahi Shirazi, A.; Parang, K. Self-assembly of peptides to nanostructures. Org. Biomol. Chem. 2014, 12, 3544–3561. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, W.; Chen, C.; Wang, J.; Zhang, L.; Xu, H. Rational design and self-assembly of short amphiphilic peptides and applications. Curr. Opin. Colloid Interface Sci. 2018, 35, 112–123. [Google Scholar] [CrossRef]
- Singh, N.; Kumar, M.; Miravet, J.F.; Ulijn, R.V.; Escuder, B. Peptide-Based Molecular Hydrogels as Supramolecular Protein Mimics. Chemistry 2017, 23, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Aumiller, W.M., Jr.; Pir Cakmak, F.; Davis, B.W.; Keating, C.D. RNA-Based Coacervates as a Model for Membraneless Organelles: Formation, Properties, and Interfacial Liposome Assembly. Langmuir 2016, 32, 10042–10053. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.Y.; Keyes, C.; Duhamel, J.; Chen, P. Concentration Effect on the Aggregation of a Self-Assembling Oligopeptide. Biophys. J. 2003, 85, 537–548. [Google Scholar] [CrossRef]
- Koga, S.; Williams, D.S.; Perriman, A.W.; Mann, S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 2011, 3, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; He, L.; Yin, Y. Magnetic field guided colloidal assembly. Mater. Today 2013, 16, 110–116. [Google Scholar] [CrossRef]
- Yin, Y.; Niu, L.; Zhu, X.; Zhao, M.; Zhang, Z.; Mann, S.; Liang, D. Non-equilibrium behaviour in coacervate-based protocells under electric-field-induced excitation. Nat. Commun. 2016, 7, 10658. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.E.; Kelley, M.J. Interaction of mineral surfaces with simple organic molecules by diffuse reflectance IR spectroscopy (DRIFT). J. Colloid Interface Sci. 2008, 322, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Hakim, S.S.; Olsson, M.H.M.; Sørensen, H.O.; Bovet, N.; Bohr, J.; Feidenhans’l, R.; Stipp, S.L.S. Interactions of the Calcite {10.4} Surface with Organic Compounds: Structure and Behaviour at Mineral—Organic Interfaces. Sci. Rep. 2017, 7, 7592. [Google Scholar] [CrossRef] [PubMed]
- Pasarín, I.S.; Yang, M.; Bovet, N.; Glyvradal, M.; Nielsen, M.M.; Bohr, J.; Feidenhans’l, R.; Stipp, S.L.S. Molecular ordering of ethanol at the calcite surface. Langmuir 2012, 28, 2545–2550. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.A.; Ernst, M.; Meier, B.H.; Suter, U.W. Structure and Molecular Dynamics of Alkane Monolayers Self-Assembled on Mica Platelets. J. Phys. Chem. B 2002, 106, 653–662. [Google Scholar] [CrossRef]
- Duffy, D.M.; Harding, J.H. Simulation of organic monolayers as templates for the nucleation of calcite crystals. Langmuir 2004, 20, 7630–7636. [Google Scholar] [CrossRef] [PubMed]
- Sand, K.K.; Yang, M.; Makovicky, E.; Cooke, D.J.; Hassenkam, T.; Bechgaard, K.; Stipp, S.L.S. Binding of ethanol on calcite: The role of the OH bond and its relevance to biomineralization. Langmuir 2010, 26, 15239–15247. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.J.; Gray, R.J.; Sand, K.K.; Stipp, S.L.S.; Elliott, J.A. Interaction of ethanol and water with the {1014} surface of calcite. Langmuir 2010, 26, 14520–14529. [Google Scholar] [CrossRef] [PubMed]
- Yaalon, D.H. Mineral Composition of the Average Shale. Clay Miner. 1962, 5, 31–36. [Google Scholar] [CrossRef]
- Banerjee, S.; Abdulsattar, Z.R.; Agim, K.; Lane, R.H.; Hascakir, B. Mechanism of polymer adsorption on shale surfaces: Effect of polymer type and presence of monovalent and divalent salts. Petroleum 2017, 3, 384–390. [Google Scholar] [CrossRef]
- Carter, P.W. Adsorption of amino acid-containing organic matter by calcite and quartz. Geochim. Cosmochim. Acta 1978, 42, 1239–1242. [Google Scholar] [CrossRef]
- Lagaly, G.; Barrer, R.M.; Goulding, K. Clay-Organic Interactions. Philos. Trans. Royal Soc. A Math. Phys. Eng. Sci. 1984, 311, 315–332. [Google Scholar] [CrossRef]
- Mao, C. The emergence of complexity: Lessons from DNA. PLoS Biol. 2004, 2, e431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C.; Breslauer, K.J.; Remeta, D.P.; Minetti, C.A. What drives proteins into the major or minor grooves of DNA? J. Mol. Biol. 2007, 365, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Walter, N.G.; Engelke, D.R. Ribozymes: Catalytic RNAs that cut things, make things, and do odd and useful jobs. Biologist 2002, 49, 199–203. [Google Scholar] [PubMed]
- Robertson, M.P.; Joyce, G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4, a003608. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Prywes, N.; Tam, C.P.; O’Flaherty, D.K.; Lelyveld, V.S.; Izgu, E.C.; Pal, A.; Szostak, J.W. Enhanced Nonenzymatic RNA Copying with 2-Aminoimidazole Activated Nucleotides. J. Am. Chem. Soc. 2017, 139, 1810–1813. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012, 3, 2. [Google Scholar] [CrossRef]
- Sowerby, S.J.; Heckl, W.M. The role of self-assembled monolayers of the purine and pyrimidine bases in the emergence of life. Orig. Life Evol. Biosph. 1998, 28, 283–310. [Google Scholar] [CrossRef] [PubMed]
- Kan, Y.; Tan, Q.; Wu, G.; Si, W.; Chen, Y. Study of DNA adsorption on mica surfaces using a surface force apparatus. Sci. Rep. 2015, 5, 8442. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Huang, Q.-Y.; Zhang, X.-W. Interactions of DNA with Clay Minerals and Soil Colloidal Particles and Protection against Degradation by DNase. Environ. Sci. Technol. 2006, 40, 2971–2976. [Google Scholar] [CrossRef] [PubMed]
- Cleaves, H.J., II; Crapster-Pregont, E.; Jonsson, C.M.; Jonsson, C.L.; Sverjensky, D.A.; Hazen, R.A. The adsorption of short single-stranded DNA oligomers to mineral surfaces. Chemosphere 2011, 83, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Perona, J.J. DNA Structure. In Encyclopedia of Genetics; Elsevier: New York, NY, USA, 2001; pp. 572–574. ISBN 9780122270802. [Google Scholar]
- Herbert, A.; Rich, A. The Biology of Left-handed Z.-DNA. J. Biol. Chem. 1996, 271, 11595–11598. [Google Scholar] [CrossRef] [PubMed]
- Freund, A.M.; Bichara, M.; Fuchs, R.P. Z-DNA-forming sequences are spontaneous deletion hot spots. Proc. Natl. Acad. Sci. USA 1989, 86, 7465–7469. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Christensen, L.A.; Vasquez, K.M. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc. Natl. Acad. Sci. USA 2006, 103, 2677–2682. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; Zhang, S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003, 4, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, Fifth Edition; W. H. Freeman: New York, NY, USA, 2002; ISBN 9780716730514. [Google Scholar]
- Swadling, J.B.; Coveney, P.V.; Christopher Greenwell, H. Stability of free and mineral-protected nucleic acids: Implications for the RNA world. Geochim. Cosmochim. Acta 2012, 83, 360–378. [Google Scholar] [CrossRef]
- Riazance, J.H.; Baase, W.A.; Johnson, W.C., Jr.; Hall, K.; Cruz, P.; Tinoco, I., Jr. Evidence for Z-form RNA by vacuum UV circular dichroism. Nucleic Acids Res. 1985, 13, 4983–4989. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.W.; Adamiak, R.W.; Tinoco, I., Jr. Z-RNA: The solution NMR structure of r(CGCGCG). Biopolymers 1990, 29, 109–122. [Google Scholar] [CrossRef] [PubMed]
- D’Ascenzo, L.; Leonarski, F.; Vicens, Q.; Auffinger, P. “Z-DNA like” fragments in RNA: A recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016, 44, 5944–5956. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, B.D.; Pal, A.; Del Frate, F.; Topkar, V.V.; Szostak, J.W. Replacing uridine with 2-thiouridine enhances the rate and fidelity of nonenzymatic RNA primer extension. J. Am. Chem. Soc. 2015, 137, 2769–2775. [Google Scholar] [CrossRef] [PubMed]
- Franchi, M.; Ferris, J.P.; Gallori, E. Cations as mediators of the adsorption of nucleic acids on clay surfaces in prebiotic environments. Orig. Life Evol. Biosph. 2003, 33, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G.; Laney, D.E. DNA binding to mica correlates with cationic radius: Assay by atomic force microscopy. Biophys. J. 1996, 70, 1933–1939. [Google Scholar] [CrossRef]
- Moore, E.K.; Hao, J.; Prabhu, A.; Zhong, H.; Jelen, B.I.; Meyer, M.; Hazen, R.M.; Falkowski, P.G. Geological and Chemical Factors that Impacted the Biological Utilization of Cobalt in the Archean Eon. J. Geophys. Res. Biogeosci. 2018, 123, 743–759. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. A 1976, 32, 751–767. [Google Scholar] [CrossRef]
- Crowe, S.A.; Jones, C.; Katsev, S.; Magen, C.; O’Neill, A.H.; Sturm, A.; Canfield, D.E.; Haffner, G.D.; Mucci, A.; Sundby, B.; et al. Photoferrotrophs thrive in an Archean Ocean analogue. Proc. Natl. Acad. Sci. USA 2008, 105, 15938–15943. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hyeon Ko, S.; Zhang, C.; Ribbe, A.E.; Mao, C. Surface-mediated DNA self-assembly. J. Am. Chem. Soc. 2009, 131, 13248–13249. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Rothemund, P.W.K. Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nat. Commun. 2014, 5, 4889. [Google Scholar] [CrossRef] [PubMed]
- Perrault, S.D.; Shih, W.M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The Beauty and Utility of DNA Origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Akbari, E.; Mollica, M.Y.; Lucas, C.R.; Bushman, S.M.; Patton, R.A.; Shahhosseini, M.; Song, J.W.; Castro, C.E. Engineering Cell Surface Function with DNA Origami. Adv. Mater. 2017, 29. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Qi, X.; Myhrvold, C.; Wang, B.; Dai, M.; Jiang, S.; Bates, M.; Liu, Y.; An, B.; Zhang, F.; et al. Single-stranded DNA and RNA origami. Science 2017, 358, eaao2648. [Google Scholar] [CrossRef] [PubMed]
- Oi, H.; Fujita, D.; Suzuki, Y.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. Programmable formation of catalytic RNA triangles and squares by assembling modular RNA enzymes. J. Biochem. 2017, 161, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Matsumura, S.; Ikawa, Y. Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology 2017, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, G.; Christian, A.; Inoue, T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G. A century of Alzheimer’s disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. The Amyloid Beta Peptide: A Chemist’s Perspective. Role in Alzheimer’s and Fibrillization. Chem. Rev. 2012, 112, 5147–5192. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.T.; Vieira, M.N.N.; De Felice, F.G. Soluble protein oligomers as emerging toxins in Alzheimer’s and other amyloid diseases. IUBMB Life 2007, 59, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Eisele, Y.S.; Monteiro, C.; Fearns, C.; Encalada, S.E.; Wiseman, R.L.; Powers, E.T.; Kelly, J.W. Targeting protein aggregation for the treatment of degenerative diseases. Nat. Rev. Drug Discov. 2015, 14, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Ow, S.-Y.; Dunstan, D.E. A brief overview of amyloids and Alzheimer’s disease. Protein Sci. 2014, 23, 1315–1331. [Google Scholar] [CrossRef] [PubMed]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Frederix, P.W.J.; Scott, G.G.; Abul-Haija, Y.M.; Kalafatovic, D.; Pappas, C.G.; Javid, N.; Hunt, N.T.; Ulijn, R.V.; Tuttle, T. Exploring the sequence space for (tri-)peptide self-assembly to design and discover new hydrogels. Nat. Chem. 2014, 7, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappas, C.G.; Shafi, R.; Sasselli, I.R.; Siccardi, H.; Wang, T.; Narang, V.; Abzalimov, R.; Wijerathne, N.; Ulijn, R.V. Dynamic peptide libraries for the discovery of supramolecular nanomaterials. Nat. Nanotechnol. 2016, 11, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Adler-Abramovich, L.; Gazit, E. The physical properties of supramolecular peptide assemblies: From building block association to technological applications. Chem. Soc. Rev. 2014, 43, 6881–6893. [Google Scholar] [CrossRef] [PubMed]
- Schnaider, L.; Brahmachari, S.; Schmidt, N.W.; Mensa, B.; Shaham-Niv, S.; Bychenko, D.; Adler-Abramovich, L.; Shimon, L.J.W.; Kolusheva, S.; DeGrado, W.F.; et al. Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat. Commun. 2017, 8, 1365. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Makam, P.; Aizen, R.; Gazit, E. Self-assembling peptide semiconductors. Science 2017, 358, eaam9756. [Google Scholar] [CrossRef] [PubMed]
- Galit Fichman, E.G. Self-assembly of short peptides to form hydrogels: Design of building blocks, physical properties and technological applications. Acta Biomater. 2014, 10, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Plankensteiner, K.; Reiner, H.; Schranz, B.; Rode, B.M. Prebiotic Formation of Amino Acids in a Neutral Atmosphere by Electric Discharge. Angew. Chem. Int. Ed. 2004, 43, 1886–1888. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L. The atmosphere of the primitive Earth and the prebiotic synthesis of amino acids. Orig. Life 1974, 5, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Sephton, M.A. Organic compounds in carbonaceous meteorites. Nat. Prod. Rep. 2002, 19, 292–311. [Google Scholar] [CrossRef] [PubMed]
- Hennet, R.J.-C.; Holm, N.G.; Engel, M.H. Abiotic synthesis of amino acids under hydrothermal conditions and the origin of life: A perpetual phenomenon? Naturwissenschaften 1992, 79, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Marshall, W.L. Hydrothermal synthesis of amino acids. Geochim. Cosmochim. Acta 1994, 58, 2099–2106. [Google Scholar] [CrossRef]
- Leman, L.J.; Huang, Z.-Z.; Reza Ghadiri, M. Peptide Bond Formation in Water Mediated by Carbon Disulfide. Astrobiology 2015, 15, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Leman, L. Carbonyl Sulfide-Mediated Prebiotic Formation of Peptides. Science 2004, 306, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Erastova, V.; Degiacomi, M.T.; Fraser, D.G.; Greenwell, H.C. Mineral surface chemistry control for origin of prebiotic peptides. Nat. Commun. 2017, 8, 2033. [Google Scholar] [CrossRef] [PubMed]
- Martra, G.; Deiana, C.; Sakhno, Y.; Barberis, I.; Fabbiani, M.; Pazzi, M.; Vincenti, M. The formation and self-assembly of long prebiotic oligomers produced by the condensation of unactivated amino acids on oxide surfaces. Angew. Chem. Int. Ed. Engl. 2014, 53, 4671–4674. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-G.; Li, H.; Huynh, T.; Zhang, F.; Xia, Z.; Zhang, Y.; Zhou, R. Molecular mechanism of surface-assisted epitaxial self-assembly of amyloid-like peptides. ACS Nano 2012, 6, 9276–9282. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, F.; Zhang, Y.; Ye, M.; Zhou, B.; Tang, Y.-Z.; Yang, H.-J.; Xie, M.-Y.; Chen, S.-F.; He, J.-H.; et al. Peptide diffusion and self-assembly in ambient water nanofilm on mica surface. J. Phys. Chem. B 2009, 113, 8795–8799. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Souillac, P.O.; Ionescu-Zanetti, C.; Carter, S.A.; Fink, A.L. Surface-catalyzed amyloid fibril formation. J. Biol. Chem. 2002, 277, 50914–50922. [Google Scholar] [CrossRef] [PubMed]
- Rout, S.K.; Friedmann, M.P.; Riek, R.; Greenwald, J. A prebiotic template-directed peptide synthesis based on amyloids. Nat. Commun. 2018, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Narimatsu, T.; Tsuchiya, S.; Tanaka, T.; Li, P.; Hayamizu, Y. Water stability of self-assembled peptide nanostructures for sequential formation of two-dimensional interstitial patterns on layered materials. RSC Adv. 2016, 6, 96889–96897. [Google Scholar] [CrossRef]
- Sadler, K.; Tam, J.P. Peptide dendrimers: Applications and synthesis. Rev. Mol. Biotechnol. 2002, 90, 195–229. [Google Scholar] [CrossRef]
- Holliday, G.L.; Mitchell, J.B.O.; Thornton, J.M. Understanding the functional roles of amino acid residues in enzyme catalysis. J. Mol. Biol. 2009, 390, 560–577. [Google Scholar] [CrossRef] [PubMed]
- Dodson, G. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Lagnoux, D.; Delort, E.; Douat-Casassus, C.; Esposito, A.; Reymond, J.-L. Synthesis and esterolytic activity of catalytic peptide dendrimers. Chemistry 2004, 10, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-S.; Lin, J.; Liu, Y.; Huang, P.; Jin, A.; Chen, X. Self-assembly mechanisms of nanofibers from peptide amphiphiles in solution and on substrate surfaces. Nanoscale 2016, 8, 14814–14820. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Luo, J.-B.; Yang, S.-T.; Zhou, Q.-H. Template-directed self-assembly of a designed amphiphilic hexapeptide on mica surface. Colloid Polym. Sci. 2013, 291, 2263–2270. [Google Scholar] [CrossRef]
- Cui, H.; Webber, M.J.; Stupp, S.I. Self-Assembly of Peptide Amphiphiles: From Molecules to Nanostructures to Biomaterials. Biopolymers 2010, 94, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Mikhalevich, V.; Craciun, I.; Kyropoulou, M.; Palivan, C.G.; Meier, W. Amphiphilic Peptide Self-Assembly: Expansion to Hybrid Materials. Biomacromolecules 2017, 18, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Self-assembly of amphiphilic peptides. Soft Matter 2011, 7, 4122–4138. [Google Scholar] [CrossRef]
- Vauthey, S.; Santoso, S.; Gong, H.; Watson, N.; Zhang, S. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc. Natl. Acad. Sci. USA 2002, 99, 5355–5360. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, C.; Szostak, J.W. Controlled growth of filamentous fatty acid vesicles under flow. Langmuir 2014, 30, 14916–14925. [Google Scholar] [CrossRef] [PubMed]
- Adamala, K.; Szostak, J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science 2013, 342, 1098–1100. [Google Scholar] [CrossRef] [PubMed]
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 2001, 294, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.; Moulton, K.; Krevor, S. Pore-scale heterogeneity in the mineral distribution and reactive surface area of porous rocks. Chem. Geol. 2015, 411, 260–273. [Google Scholar] [CrossRef]
- Jonker, A.M.; Dennis, W.P.; van Hest, J.C.M. Peptide- and Protein-Based Hydrogels. Chem. Mater. 2012, 24, 759–773. [Google Scholar] [CrossRef]
- Lei, Z.; Wang, Q.; Sun, S.; Zhu, W.; Wu, P. A Bioinspired Mineral Hydrogel as a Self-Healable, Mechanically Adaptable Ionic Skin for Highly Sensitive Pressure Sensing. Adv. Mater. 2017, 29, 1700321. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Lee, M.; Taraban, M.; Yu, Y.B. Diffusion of small molecules inside a peptide hydrogel. Chem. Commun. 2011, 47, 10455–10457. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mooney, D.J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Gállego, I.; Laughlin, B.; Grover, M.A.; Hud, N.V. A viscous solvent enables information transfer from gene-length nucleic acids in a model prebiotic replication cycle. Nat. Chem. 2016, 9, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Caliari, S.R.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Gungormus, M.; Branco, M.; Fong, H.; Schneider, J.P.; Tamerler, C.; Sarikaya, M. Self assembled bi-functional peptide hydrogels with biomineralization-directing peptides. Biomaterials 2010, 31, 7266–7274. [Google Scholar] [CrossRef] [PubMed]
- Kasotakis, E.; Mossou, E.; Adler-Abramovich, L.; Mitchell, E.P.; Forsyth, V.T.; Gazit, E.; Mitraki, A. Design of metal-binding sites onto self-assembled peptide fibrils. Biopolymers 2009, 92, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.Z.; Hentrich, C.; Szostak, J.W. Rapid RNA exchange in aqueous two-phase system and coacervate droplets. Orig. Life Evol. Biosph. 2014, 44, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aumiller, W.M., Jr.; Keating, C.D. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 2016, 8, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Nakamura, T.; Watanabe, K.; Noguchi, T.; Minamihata, K.; Kamiya, N.; Kimizuka, N. Self-assembly of Ni-NTA-modified β-annulus peptides into artificial viral capsids and encapsulation of His-tagged proteins. Org. Biomol. Chem. 2016, 14, 7869–7874. [Google Scholar] [CrossRef] [PubMed]
- Rufo, C.M.; Moroz, Y.S.; Moroz, O.V.; Stöhr, J.; Smith, T.A.; Hu, X.; DeGrado, W.F.; Korendovych, I.V. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 2014, 6, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Al-Garawi, Z.S.; McIntosh, B.A.; Neill-Hall, D.; Hatimy, A.A.; Sweet, S.M.; Bagley, M.C.; Serpell, L.C. The amyloid architecture provides a scaffold for enzyme-like catalysts. Nanoscale 2017, 9, 10773–10783. [Google Scholar] [CrossRef] [PubMed]
- Heuer, A.; Fink, D.; Laraia, V.; Arias, J.; Calvert, P.; Kendall, K.; Messing, G.; Blackwell, J.; Rieke, P.; Thompson, D.; et al. Innovative materials processing strategies: A biomimetic approach. Science 1992, 255, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Addadi, L.; Weiner, S. Biomineralization: Mineral formation by organisms. Phys. Scr. 2014, 89, 098003. [Google Scholar] [CrossRef]
- Dhami, N.K.; Reddy, M.S.; Mukherjee, A. Biomineralization of calcium carbonates and their engineered applications: A review. Front. Microbiol. 2013, 4, 314. [Google Scholar] [CrossRef] [PubMed]
- Kirschvink, J.L. Magnetite biomineralization and geomagnetic sensitivity in higher animals: An update and recommendations for future study. Bioelectromagnetics 1989, 10, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Tambutté, S.; Holcomb, M.; Ferrier-Pagès, C.; Reynaud, S.; Tambutté, É.; Zoccola, D.; Allemand, D. Coral biomineralization: From the gene to the environment. J. Exp. Mar. Biol. Ecol. 2011, 408, 58–78. [Google Scholar] [CrossRef]
- Fablet, R.; Pecquerie, L.; de Pontual, H.; Høie, H.; Millner, R.; Mosegaard, H.; Kooijman, S.A.L.M. Shedding light on fish otolith biomineralization using a bioenergetic approach. PLoS ONE 2011, 6, e27055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selenska-Pobell, S.; Merroun, M. Accumulation of Heavy Metals by Micro-organisms: Biomineralization and Nanocluster Formation. In Prokaryotic Cell Wall Compounds; Springer: Berlin, Germany, 2010; pp. 483–500. [Google Scholar]
- Pacton, M.; Wacey, D.; Corinaldesi, C.; Tangherlini, M.; Kilburn, M.R.; Gorin, G.E.; Danovaro, R.; Vasconcelos, C. Viruses as new agents of organomineralization in the geological record. Nat. Commun. 2014, 5, 4298. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Paterson, G.A.; Zhu, Q.; Wang, Y.; Kopylova, E.; Li, Y.; Knight, R.; Bazylinski, D.A.; Zhu, R.; Kirschvink, J.L.; et al. Origin of microbial biomineralization and magnetotaxis during the Archean. Proc. Natl. Acad. Sci. USA 2017, 114, 2171–2176. [Google Scholar] [CrossRef] [PubMed]
- Butardo, V.M.; Sreenivasulu, N. Tailoring Grain Storage Reserves for a Healthier Rice Diet and its Comparative Status with Other Cereals. Int. Rev. Cell Mol. Biol. 2016, 323, 31–70. [Google Scholar] [PubMed]
- Adeva-Andany, M.M.; González-Lucán, M.; Donapetry-García, C.; Fernández-Fernández, C.; Ameneiros-Rodríguez, E. Glycogen metabolism in humans. BBA Clin. 2016, 5, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Elieh-Ali-Komi, D.; Hamblin, M.R. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int. J. Adv. Res. (Indore) 2016, 4, 411–427. [Google Scholar]
- Hansma, H.G. Possible origin of life between mica sheets: Does life imitate mica? J. Biomol. Struct. Dyn. 2013, 31, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Nidhin, M.; Sreeram, K.J.; Nair, B.U. Polysaccharide films as templates in the synthesis of hematite nanostructures with special properties. Appl. Surf. Sci. 2012, 258, 5179–5184. [Google Scholar] [CrossRef]
- Allen, C.C.; Westall, F.; Schelble, R.T. Importance of a martian hematite site for astrobiology. Astrobiology 2001, 1, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Kinoshita, T.; Nagata, K.; Higuchi, M. Mineralization of Calcium Carbonate on Multifunctional Peptide Assembly Acting as Mineral Source Supplier and Template. Langmuir 2016, 32, 9351–9359. [Google Scholar] [CrossRef] [PubMed]
- Nonoyama, T.; Tanaka, M.; Kinoshita, T.; Nagata, F.; Sato, K.; Kato, K. Morphology control of calcium phosphate by mineralization on the β-sheet peptide template. Chem. Commun. 2010, 46, 6983. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.; Radvar, E.; Shi, Y.; Borges, J.; Pirraco, R.P.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Mata, Á.; Azevedo, H.S. Nanostructured interfacial self-assembled peptide–polymer membranes for enhanced mineralization and cell adhesion. Nanoscale 2017, 9, 13670–13682. [Google Scholar] [CrossRef] [PubMed]
- Elham, R.; Remzi, B.C.; Helena, A. Self-assembled peptide-polymer hydrogels as a 3D nanofiber substrate for biomimetic hydroxyapatite mineralization. Front. Bioeng. Biotechnol. 2016, 4. [Google Scholar] [CrossRef]
- Li, Q.-L.; Ning, T.-Y.; Cao, Y.; Zhang, W.-B.; Mei, M.L.; Chu, C.H. A novel self-assembled oligopeptide amphiphile for biomimetic mineralization of enamel. BMC Biotechnol. 2014, 14, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Ouyang, Z.; Ren, Z.; Li, J.; Zhang, P.; Wei, G.; Su, Z. Self-assembled peptide nanofibers on graphene oxide as a novel nanohybrid for biomimetic mineralization of hydroxyapatite. Carbon N. Y. 2015, 89, 20–30. [Google Scholar] [CrossRef]
- Chan, M.; Cleaves, H.J., II; Boston, P. What Would Earth Be Like Without Life? EOS 2018, 99. [Google Scholar] [CrossRef]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gillams, R.J.; Jia, T.Z. Mineral Surface-Templated Self-Assembling Systems: Case Studies from Nanoscience and Surface Science towards Origins of Life Research. Life 2018, 8, 10. https://doi.org/10.3390/life8020010
Gillams RJ, Jia TZ. Mineral Surface-Templated Self-Assembling Systems: Case Studies from Nanoscience and Surface Science towards Origins of Life Research. Life. 2018; 8(2):10. https://doi.org/10.3390/life8020010
Chicago/Turabian StyleGillams, Richard J., and Tony Z. Jia. 2018. "Mineral Surface-Templated Self-Assembling Systems: Case Studies from Nanoscience and Surface Science towards Origins of Life Research" Life 8, no. 2: 10. https://doi.org/10.3390/life8020010
APA StyleGillams, R. J., & Jia, T. Z. (2018). Mineral Surface-Templated Self-Assembling Systems: Case Studies from Nanoscience and Surface Science towards Origins of Life Research. Life, 8(2), 10. https://doi.org/10.3390/life8020010