Performance of Targeted Library Preparation Solutions for SARS-CoV-2 Whole Genome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Controls Generation
2.3. Calculation of Ct for Samples and Controls
2.4. NGS Library Preparation
2.4.1. Library Preparation—1st Attempt NEB+TWIST (NEB+TWIST1)
2.4.2. Library Preparation—2nd Attempt NEB+TWIST (NEB+TWIST2)
2.4.3. Library Preparation—Illumina
2.4.4. Library Preparation—Paragon
2.5. Sequencing
2.6. Reference Mapping and Bioinformatic Analysis
3. Results and Discussion
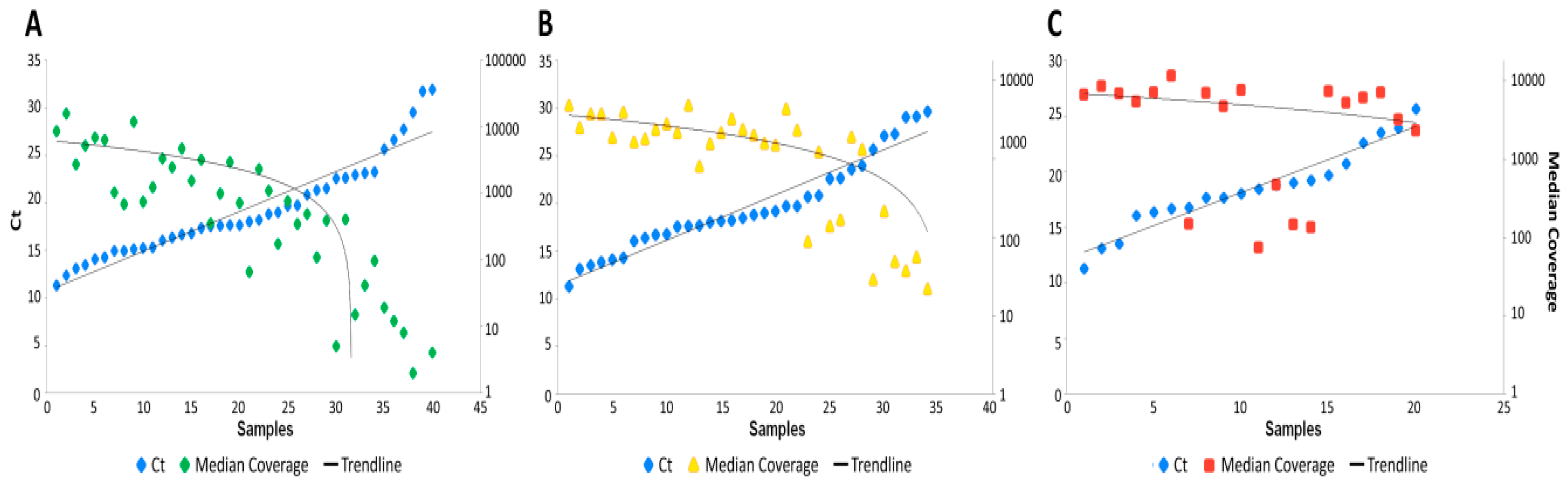
3.1. NEB+Twist Workflow
3.2. Illumina Workflow
3.3. Paragon Workflow
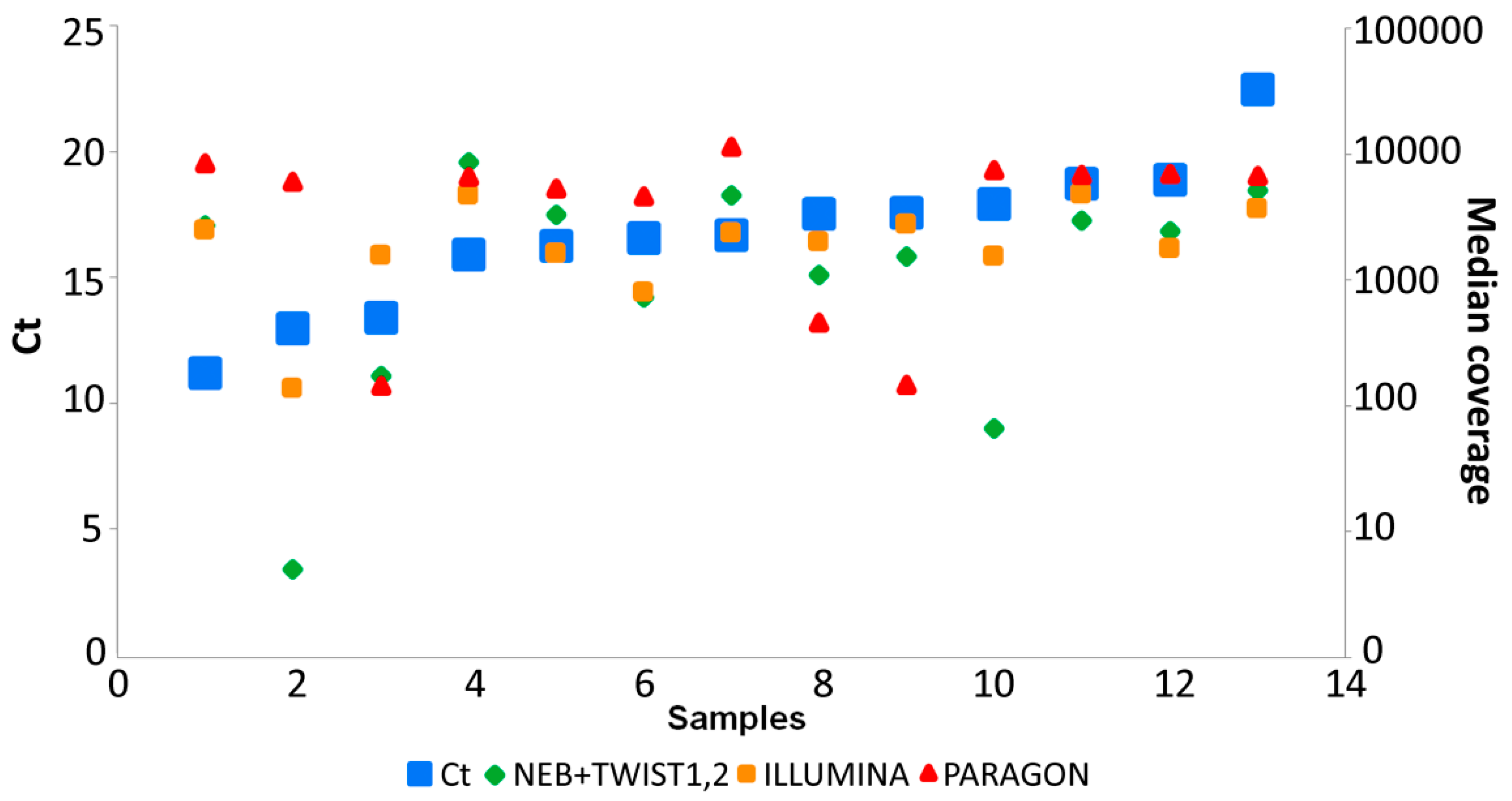
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Park, S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19). Clin. Exp. Pediatr. 2020, 63, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus that Causes a Global Threat. Int. J. Boil. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alm, E.; Broberg, E.K.; Connor, T.R.; Hodcroft, E.B.; Komissarov, A.B.; Maurer-Stroh, S.; Melidou, A.; Neher, R.A.; O’Toole, A.; Pereyaslov, D.; et al. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance 2020, 25, 2001410. [Google Scholar] [CrossRef] [PubMed]
- Alcoba-Florez, J.; Gil-Campesino, H.; De Artola, D.G.-M.; González-Montelongo, R.; Valenzuela-Fernández, A.; Ciuffreda, L.; Flores, C. Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int. J. Infect. Dis. 2020, 99, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Kriegova, E.; Fillerova, R.; Kvapil, P. Direct-RT-qPCR Detection of SARS-CoV-2 without RNA Extraction as Part of a COVID-19 Testing Strategy: From Sample to Result in One Hour. Diagnostics 2020, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.; Boshier, F.A.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020, 83, 104351. [Google Scholar] [CrossRef] [PubMed]
- Artesi, M.; Bontems, S.; Göbbels, P.; Franckh, M.; Maes, P.; Boreux, R.; Meex, C.; Melin, P.; Hayette, M.-P.; Bours, V.; et al. A recurrent mutation at position 26,340 of SARS-CoV-2 is associated with failure of the E-gene qRT-PCR utilized in a commercial dual-target diagnostic assay. J. Clin. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.D.; Muñoz, M.; Hernández, C.; Flórez, C.; Gomez, S.; Turca, A.; Pardo, L.; Barros, E.C.; Mondolfi, A.E.P. Genetic Diversity Among SARS-CoV2 Strains in South America may Impact Performance of Molecular Detection. Pathogens 2020, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef] [PubMed]
- Samorodnitsky, E.; Jewell, B.M.; Hagopian, R.; Miya, J.; Wing, M.R.; Lyon, E.; Damodaran, S.; Bhatt, D.; Reeser, J.W.; Datta, J.; et al. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Hum. Mutat. 2015, 36, 903–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, S.S.; Meissner, B.; Chavez, E.; Ben-Neriah, S.; Ennishi, D.; Jones, M.R.; Shulha, H.P.; Chan, F.C.; Boyle, M.; Kridel, R.; et al. Assessment of Capture and Amplicon-Based Approaches for the Development of a Targeted Next-Generation Sequencing Pipeline to Personalize Lymphoma Management. J. Mol. Diagn. 2018, 20, 203–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Center for Immunization and Respiratory Diseases (NCIRD). Division of Viral Diseases. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (accessed on 15 September 2020).
- ‘FastQC: A Quality Control Tool for High Throughput Sequence Data—ScienceOpen’. n.d. Available online: https://www.scienceopen.com/document?vid=de674375-ab83-4595-afa9-4c8aa9e4e736 (accessed on 10 July 2020).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ‘Picard Tools—By Broad Institute’. n.d. Available online: https://broadinstitute.github.io/picard/ (accessed on 10 July 2020).
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]



| Position | Twist1 | Coverage | Twist2 | Coverage | Illumina | Coverage | Paragon | Coverage |
|---|---|---|---|---|---|---|---|---|
| 19065 | yes | 390 | yes | 36 | yes | 117 | yes | 66 |
| 22303 | yes | 381 | yes | 62 | yes | 166 | no | 3 |
| 26144 | yes | 217 | yes | 40 | yes | 197 | yes | 45 |
| 29749 | yes | 573 | yes | 22 | yes | 58 | no | 14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klempt, P.; Brož, P.; Kašný, M.; Novotný, A.; Kvapilová, K.; Kvapil, P. Performance of Targeted Library Preparation Solutions for SARS-CoV-2 Whole Genome Analysis. Diagnostics 2020, 10, 769. https://doi.org/10.3390/diagnostics10100769
Klempt P, Brož P, Kašný M, Novotný A, Kvapilová K, Kvapil P. Performance of Targeted Library Preparation Solutions for SARS-CoV-2 Whole Genome Analysis. Diagnostics. 2020; 10(10):769. https://doi.org/10.3390/diagnostics10100769
Chicago/Turabian StyleKlempt, Petr, Petr Brož, Martin Kašný, Adam Novotný, Kateřina Kvapilová, and Petr Kvapil. 2020. "Performance of Targeted Library Preparation Solutions for SARS-CoV-2 Whole Genome Analysis" Diagnostics 10, no. 10: 769. https://doi.org/10.3390/diagnostics10100769





