MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples Collection
2.2. Nucleic Acid Extraction
2.3. ASqPCR Assays for MYD88L265P Detection
2.4. ddPCR Assays for MYD88L265P Detection
2.5. Statistical Analysis
3. Results
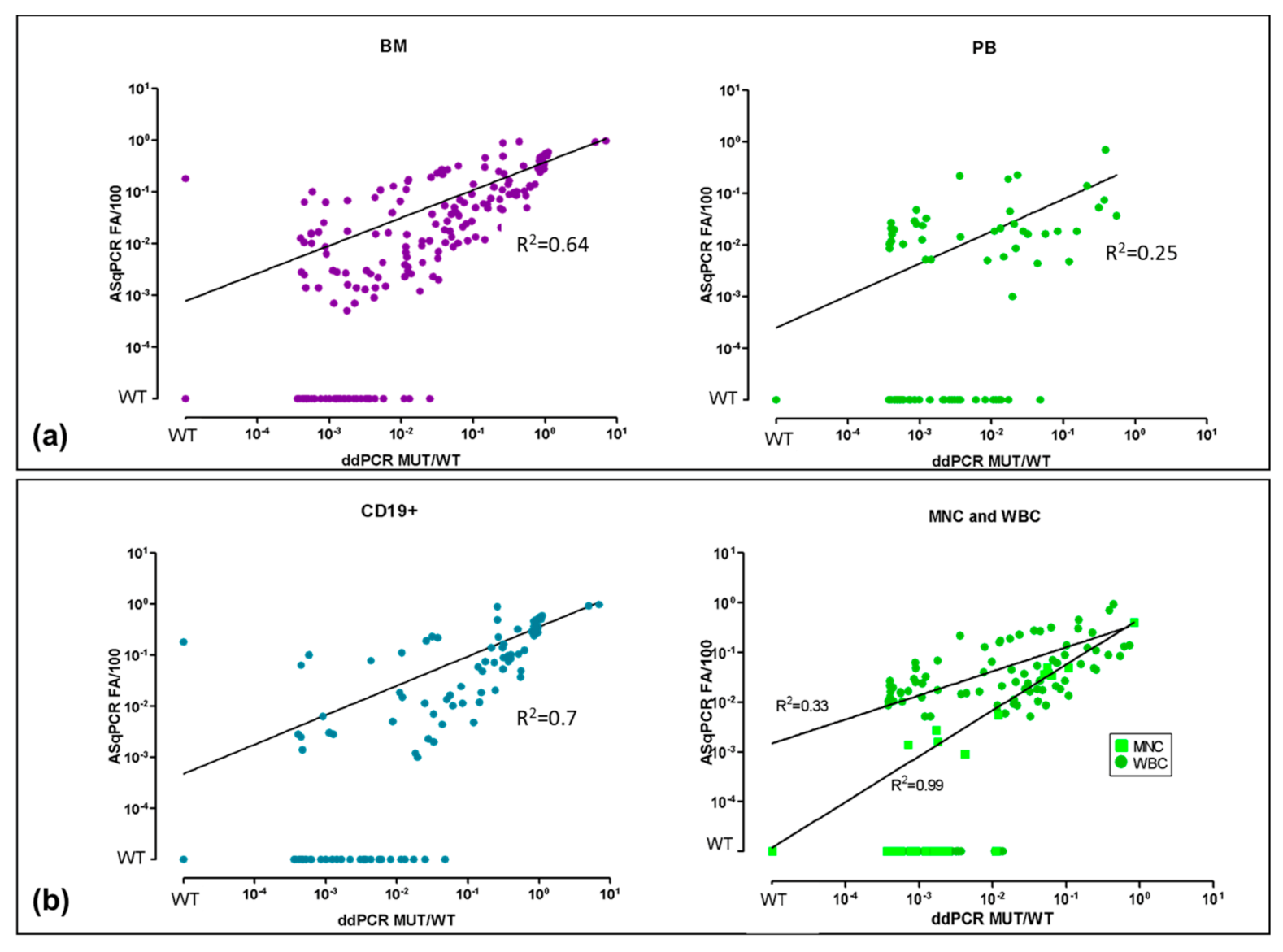
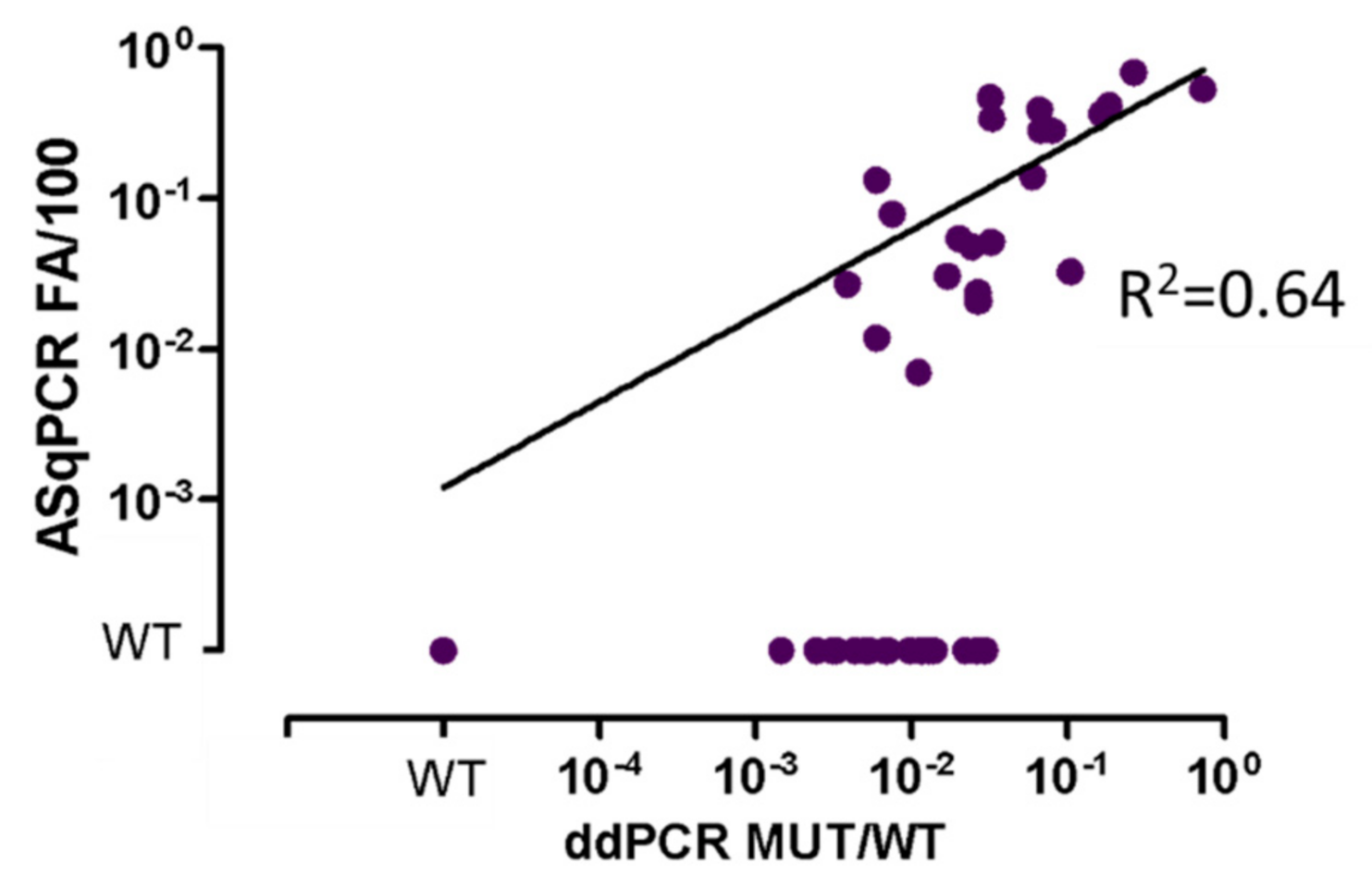
3.1. ddPCR vs. ASqPCR in Different Tissues (BM vs. PB)
3.2. ddPCR vs. ASqPCR in CD19+ Sorted vs. Unsorted Samples
3.3. ddPCR vs. ASqPCR in WM Compared to IgM-MGUS
3.4. ddPCR vs. ASqPCR in Plasma-cfDNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Girard, L.-P.; Soekojo, C.; Ooi, M.; Poon, L.; Chng, W.-J.; Mel, S. Immunoglobulin M Paraproteinaemias. Cancers 2020, 12, 1688. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Thomson, P.J.; Nikoloutsopoulos, T.; Smith, A.J.F. IgM paraproteinaemia: Disease associations and laboratory features. Pathology 2002, 34, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Dispenzieri, A.; Kumar, S.; Larson, D.; Therneau, T.; Vincent Rajkumar, S. IgM monoclonal gammopathy of undetermined significance (MGUS) and smoldering Waldenström’s macroglobulinemia (SWM). Clin. Lymphoma Myeloma Leuk. 2011, 11, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Rajkumar, S.V. Monoclonal gammopathies of undetermined significance: A review. Immunol. Rev. 2003, 194, 112–139. [Google Scholar] [CrossRef]
- Owen, R.G.; Treon, S.P.; Al-Katib, A.; Fonseca, R.; Greipp, P.R.; McMaster, M.L.; Morra, E.; Pangalis, G.A.; San Miguel, J.F.; Branagan, A.R.; et al. Clinicopathological definition of Waldenstrom’s macroglobulinemia: Consensus Panel Recommendations from the Second International Workshop on Waldenstrom’s Macroglobulinemia. Semin. Oncol. 2003, 30, 110–115. [Google Scholar] [CrossRef]
- Gertz, M.A. Waldenström macroglobulinemia: 2021 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2021, 96, 258–269. [Google Scholar] [CrossRef]
- Paiva, B.; Montes, M.C.; García-Sanz, R.; Ocio, E.M.; Alonso, J.; de Las Heras, N.; Escalante, F.; Cuello, R.; de Coca, A.G.; Galende, J.; et al. Multiparameter flow cytometry for the identification of the Waldenström’s clone in IgM-MGUS and Waldenström’s Macroglobulinemia: New criteria for differential diagnosis and risk stratification. Leukemia 2014, 28, 166–173. [Google Scholar] [CrossRef]
- Kastritis, E.; Morel, P.; Duhamel, A.; Gavriatopoulou, M.; Kyrtsonis, M.C.; Durot, E.; Symeonidis, A.; Laribi, K.; Hatjiharissi, E.; Ysebaert, L.; et al. A revised international prognostic score system for Waldenström’s macroglobulinemia. Leukemia 2019, 33, 2654–2661. [Google Scholar] [CrossRef]
- Treon, S.P.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Sheehy, P.; Manning, R.J.; Patterson, C.J.; Tripsas, C.; et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N. Engl. J. Med. 2012, 367, 826–833. [Google Scholar] [CrossRef] [Green Version]
- Treon, S.P.; Xu, L.; Guerrera, M.L.; Jimenez, C.; Hunter, Z.R.; Liu, X.; Demos, M.; Gustine, J.; Chan, G.; Munshi, M.; et al. Genomic landscape of Waldenström macroglobulinemia and its impact on treatment strategies. J. Clin. Oncol. 2020, 38, 1198–1208. [Google Scholar] [CrossRef]
- Jiménez, C.; Sebastián, E.; Chillón, M.C.; Giraldo, P.; Mariano Hernández, J.; Escalante, F.; González-López, T.J.; Aguilera, C.; De Coca, A.G.; Murillo, I.; et al. MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenström’s macroglobulinemia. Leukemia 2013, 27, 1722–1728. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, C.; Chillón Mdel, C.; Balanzategui, A.; Puig, N.; Sebastián, E.; Alcoceba, M.; Sarasquete, M.E.; Conde, I.P.; Corral, R.; Marín, L.A.; et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Appl. Immunohistochem. Mol Morphol. 2013, 121, 2051–2058. [Google Scholar] [CrossRef]
- Landgren, L.; Staudt, O. MYD88 L265P somatic mutation in IgM MGUS. N. Engl. J. Med. 2012, 367, 2255–2257. [Google Scholar] [CrossRef]
- Poulain, S.; Roumier, C.; Decambron, A.; Renneville, A.; Herbaux, C.; Bertrand, E.; Tricot, S.; Daudignon, A.; Galiègue-Zouitina, S.; Soenen, V.; et al. MYD88 L265P mutation in Waldenstrom macroglobulinemia. Blood 2013, 121, 4504–4511. [Google Scholar] [CrossRef]
- Argentou, N.; Vassilopoulos, G.; Ioannou, M.; Germenis, A.E.; Speletas, M. Rapid detection of MYD88-L265P mutation by PCR-RFLP in B-cell lymphoproliferative disorders. Leukemia 2013, 28, 447–449. [Google Scholar] [CrossRef]
- Varettoni, M.; Arcaini, L.; Zibellini, S.; Boveri, E.; Rattotti, S.; Riboni, R.; Corso, A.; Orlandi, E.; Bonfichi, M.; Gotti, M.; et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenström’s macroglobulinemia and related lymphoid neoplasms. Blood 2013, 121, 2522–2528. [Google Scholar] [CrossRef] [Green Version]
- Treon, S.P.; Cao, Y.; Xu, L.; Yang, G.; Liu, X.H.Z. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenstrom macroglobulinemia. Blood 2014, 123, 2791–2796. [Google Scholar] [CrossRef]
- Treon, S.P.; Gustine, J.; Xu, L.; Manning, R.J.; Tsakmaklis, N.; Demos, M.; Meid, K.; Guerrera, M.L.; Munshi, M.; Chan, G.; et al. MYD88 wild-type Waldenstrom Macroglobulinaemia: Differential diagnosis, risk of histological transformation, and overall survival. Br. J. Haematol. 2018, 180, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Hunter, Z.R.; Xu, L.; Yang, G.; Zhou, Y.; Liu, X.; Cao, Y.; Manning, R.J.; Tripsas, C.; Patterson, C.J.; Sheehy, P.; et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood 2014, 123, 1637–1646. [Google Scholar] [CrossRef] [Green Version]
- Treon, S.P.; Tripsas, C.K.; Meid, K.; Warren, D.; Varma, G.; Green, R.; Argyropoulos, K.V.; Yang, G.; Cao, Y.; Xu, L.; et al. Ibrutinib in previously treated Waldenström’s macroglobulinemia. N. Engl. J. Med. 2015, 372, 1430–1440. [Google Scholar] [CrossRef] [Green Version]
- Treon, S.P.; Xu, L.; Hunter, Z. MYD88 Mutations and Response to Ibrutinib in Waldenström’s Macroglobulinemia. N. Engl. J. Med. 2015, 373, 584–586. [Google Scholar] [CrossRef]
- Bustoros, M.; Sklavenitis-Pistofidis, R.; Kapoor, P.; Liu, C.-J.; Kastritis, E.; Zanwar, S.; Fell, G.; Abeykoon, J.P.; Hornburg, K.; Neuse, C.J.; et al. Progression Risk Stratification of Asymptomatic Waldenström Macroglobulinemia. J. Clin. Oncol. 2019, 37, 1403–1411. [Google Scholar] [CrossRef]
- Kastritis, E.; Leblond, V.; Dimopoulos, M.; Kimby, E.; Staber, P.; Kersten, M.; Tedeschi, A.; Buske, C. Waldenström’s macroglobulinaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv41–iv50. [Google Scholar] [CrossRef]
- Jiménez, C.; Chillón, M.d.C.; Balanzategui, A.; Puig, N.; Sebastián, E.; Alcoceba, M.; Sarasquete, M.E.; Conde, I.P.; Corral, R.; Marín, L.A.; et al. Detection of MYD88 L265P Mutation by Real-Time Allele-Specific Oligonucleotide Polymerase Chain Reaction. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 768–773. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Yang, G.; Zhou, Y.; Cao, Y.; Liu, X.; Morra, E.; Trojani, A.; Greco, A.; Arcaini, L.; et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood 2013, 121, 2051–2058. [Google Scholar] [CrossRef]
- Xu, L.; Hunter, Z.R.; Yang, G.; Cao, Y.; Liu, X.; Manning, R.J.; Tripsas, C.K.; Chen, J.; Patterson, C.J.; Kluk, M.J.; et al. Detection of MYD88 L265P in peripheral blood of patients with Waldenström’s Macroglobulinemia and IgM monoclonal gammopathy of undetermined significance. Leukemia 2014, 28, 1698–1704. [Google Scholar] [CrossRef]
- Gustine, J.; Meid, K.; Xu, L.; Hunter, Z.R.; Castillo, J.J.; Treon, S.P. To select or not to select? The role of B-cell selection in determining the MYD88 mutation status in Waldenström Macroglobulinaemia. Br. J. Haematol. 2017, 176, 822–824. [Google Scholar] [CrossRef] [Green Version]
- Diaz, L.A.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lázaro, D.; Hernández, J.L.G.; García, A.C.; Martínez, A.C.; Mielgo-Ayuso, J. Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huggett, J.F.; Whale, A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin. Chem. 2013, 59, 1691–1693. [Google Scholar] [CrossRef] [Green Version]
- Drandi, D.; Kubiczkova-Besse, L.; Ferrero, S.; Dani, N.; Passera, R.; Mantoan, B.; Gambella, M.; Monitillo, L.; Saraci, E.; Ghione, P.; et al. Minimal Residual Disease Detection by Droplet Digital PCR in Multiple Myeloma, Mantle Cell Lymphoma, and Follicular Lymphoma. J. Mol. Diagn. 2015, 17, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Drandi, D.; Genuardi, E.; Dogliotti, I.; Ferrante, M.; Jiménez, C.; Guerrini, F.; Schirico, M.L.; Mantoan, B.; Muccio, V.; Lia, G.; et al. Highly sensitive MYD88 L265P mutation detection by droplet digital polymerase chain reaction in Waldenström macroglobulinemia. Haematologica 2018, 103, 1029–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagratuni, T.; Ntanasis-Stathopoulos, I.; Gavriatopoulou, M.; Mavrianou-Koutsoukou, N.; Liacos, C.; Patseas, D.; Kanellias, N.; Migkou, M.; Ziogas, D.C.; Eleutherakis-Papaiakovou, E.; et al. Detection of MYD88 and CXCR4 mutations in cell-free DNA of patients with IgM monoclonal gammopathies. Leukemia 2018, 32, 2617–2625. [Google Scholar] [CrossRef]
- Huggett, J.F.; Whale, A.S.; De Spiegelaere, W.; Trypsteen, W.; Nour, A.A.; Bae, Y.-K.; Benes, V.; Burke, D.; Cleveland, M.; The dMIQE Group; et al. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Dimopoulos, M.; Sanz, R.G.; Lee, H.P.; Trneny, M.; Varettoni, M.; Opat, S.; D’Sa, S.; Owen, R.G.; Cull, G.; Mulligan, S.; et al. Zanubrutinib for the treatment of MYD88 wild-type Waldenström macroglobulinemia: A substudy of the phase 3 ASPEN trial. Blood Adv. 2020, 4, 6009–6018. [Google Scholar] [CrossRef]
- Owen, R.G.; McCarthy, H.; Rule, S.; D’Sa, S.; Thomas, S.K.; Tournilhac, O.; Forconi, F.; Kersten, M.J.; Zinzani, P.L.; Iyengar, S.; et al. Acalabrutinib monotherapy in patients with Waldenström macroglobulinemia: A single-arm, multicentre, phase 2 study. Lancet Haematol. 2020, 7, e112–e121. [Google Scholar] [CrossRef]


| WM | BM DIA | BM FU | PB DIA | PB FU | PLASMA | ||
| WBC | CD19+ | WBC | WBC | CD19+ | WBC | cfDNA | |
| ddPCR | ++ | ++ | ++ | ++ | ++ | + | ++ |
| ASqPCR | + | ++ | + | + | ++ | +/− | +/− |
| IgM-MGUS | BM DIA | BM FU | PB DIA | PB FU | PLASMA | ||
| WBC | CD19+ | WBC | WBC | CD19+ | WBC | cfDNA | |
| ddPCR | ++ | ++ | ++ | + | + | + | + |
| ASqPCR | +/− | ++ | +/− | +/− | + | +/− | +/− |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrante, M.; Furlan, D.; Zibellini, S.; Borriero, M.; Candido, C.; Sahnane, N.; Uccella, S.; Genuardi, E.; Alessandria, B.; Bianchi, B.; et al. MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation. Diagnostics 2021, 11, 779. https://doi.org/10.3390/diagnostics11050779
Ferrante M, Furlan D, Zibellini S, Borriero M, Candido C, Sahnane N, Uccella S, Genuardi E, Alessandria B, Bianchi B, et al. MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation. Diagnostics. 2021; 11(5):779. https://doi.org/10.3390/diagnostics11050779
Chicago/Turabian StyleFerrante, Martina, Daniela Furlan, Silvia Zibellini, Michela Borriero, Chiara Candido, Nora Sahnane, Silvia Uccella, Elisa Genuardi, Beatrice Alessandria, Benedetta Bianchi, and et al. 2021. "MYD88L265P Detection in IgM Monoclonal Gammopathies: Methodological Considerations for Routine Implementation" Diagnostics 11, no. 5: 779. https://doi.org/10.3390/diagnostics11050779







