Identifying Mitotic Kinesins as Potential Prognostic Biomarkers in Ovarian Cancer Using Bioinformatic Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of DEGs
2.2. Gene Annotations
2.3. Expression Profiling Analysis for Mitotic KIFs
2.4. Survival Analysis
2.5. Function Enrichment Analysis
2.6. Pathway and Drug-Sensitivity Analysis
2.7. Immune Infiltration and Genetic Alterations Analysis
2.8. Statistics
3. Results
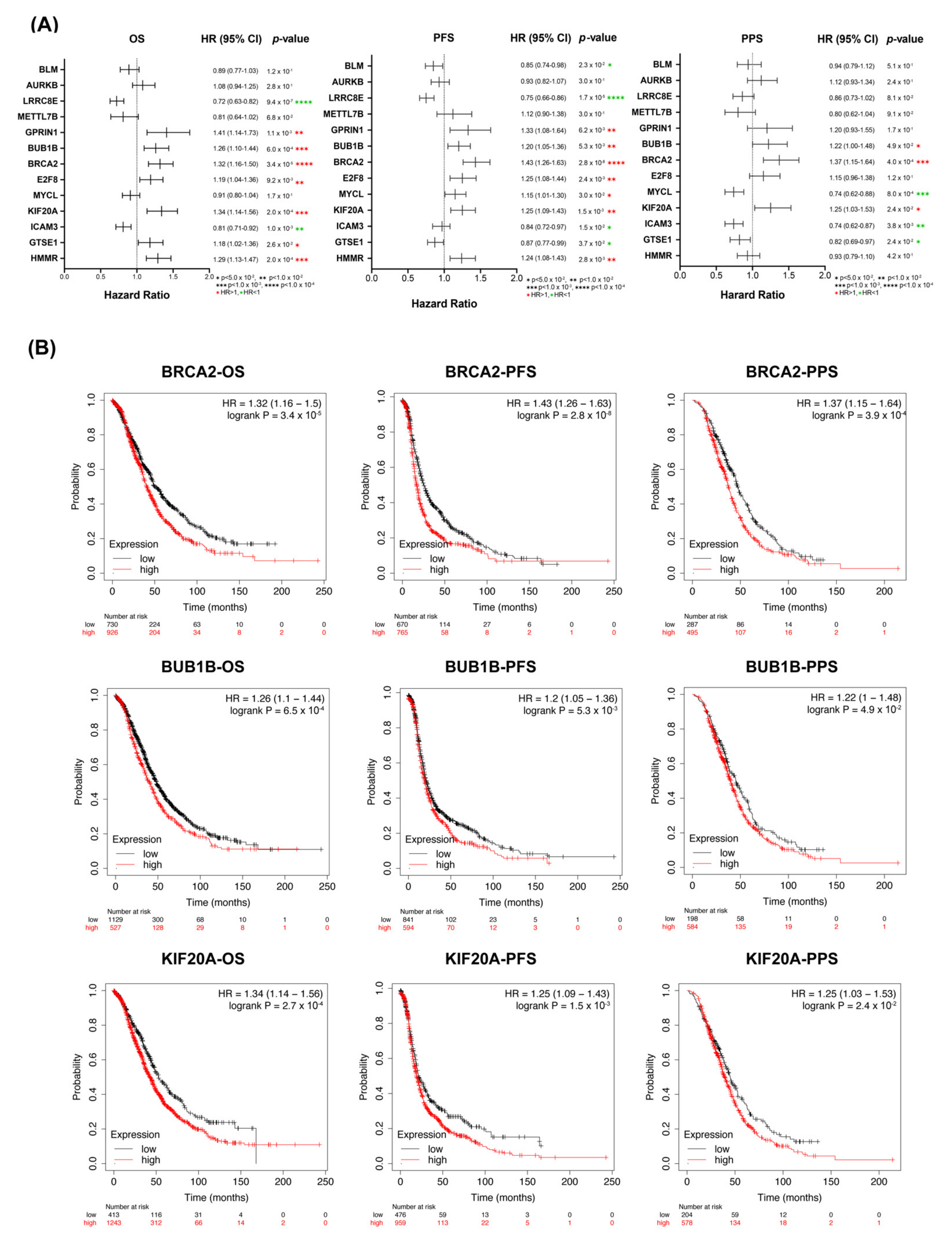
3.1. Detection of Prognostic Biomarkers
3.2. Correlation of Overexpressed of KIF20A/BRCA2/BUB1B with Poor Prognosis in OC
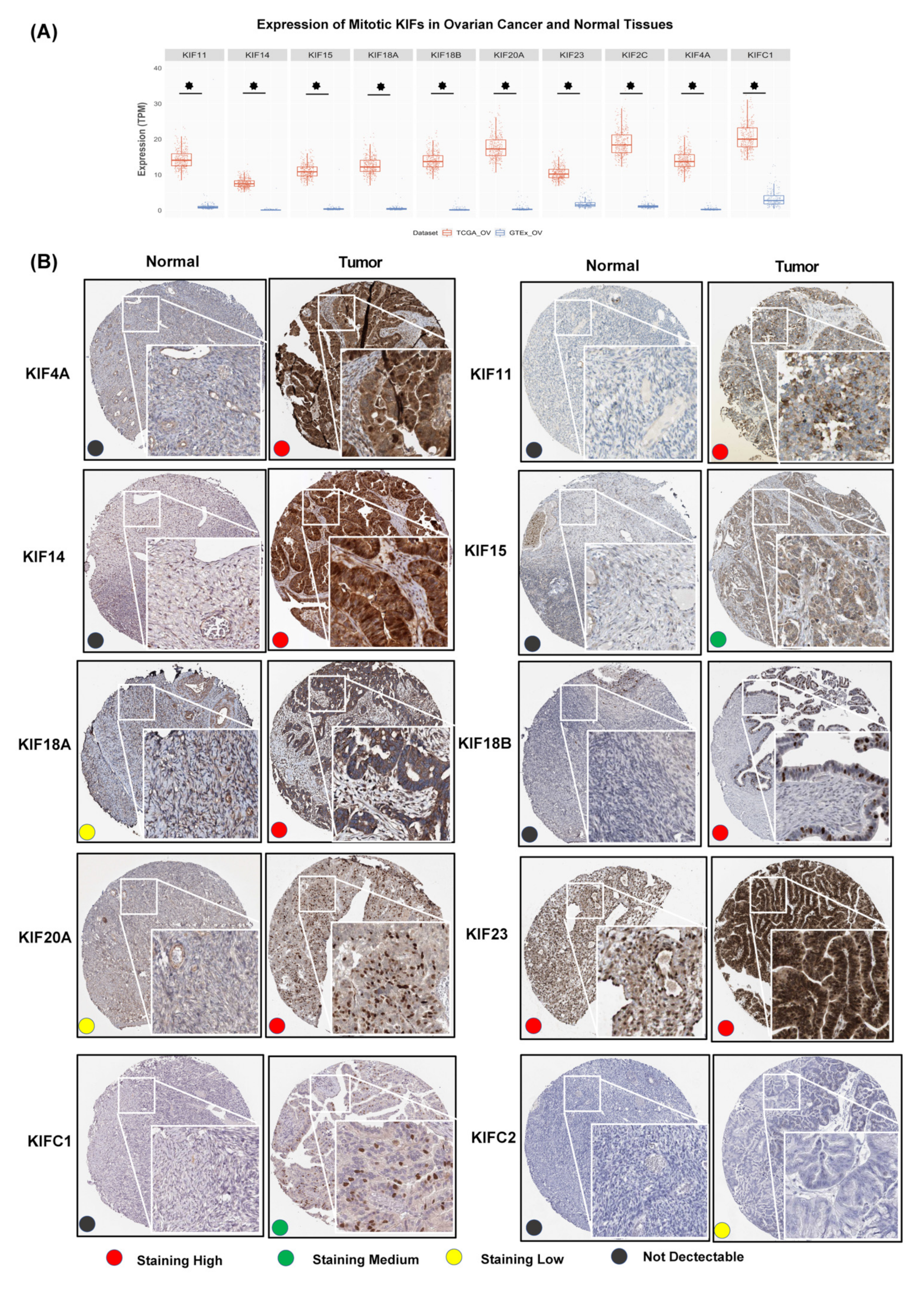
3.3. Overexpression of Further Nine Mitotic KIFs as Well as KIF20A in OC
3.4. No Correlation of Overexpressed Mitotic KIFs with Tumor-Infiltrating Lymphocytes
3.5. Correlation of Overexpressed Mitotic KIFs with High Cellular Proliferation
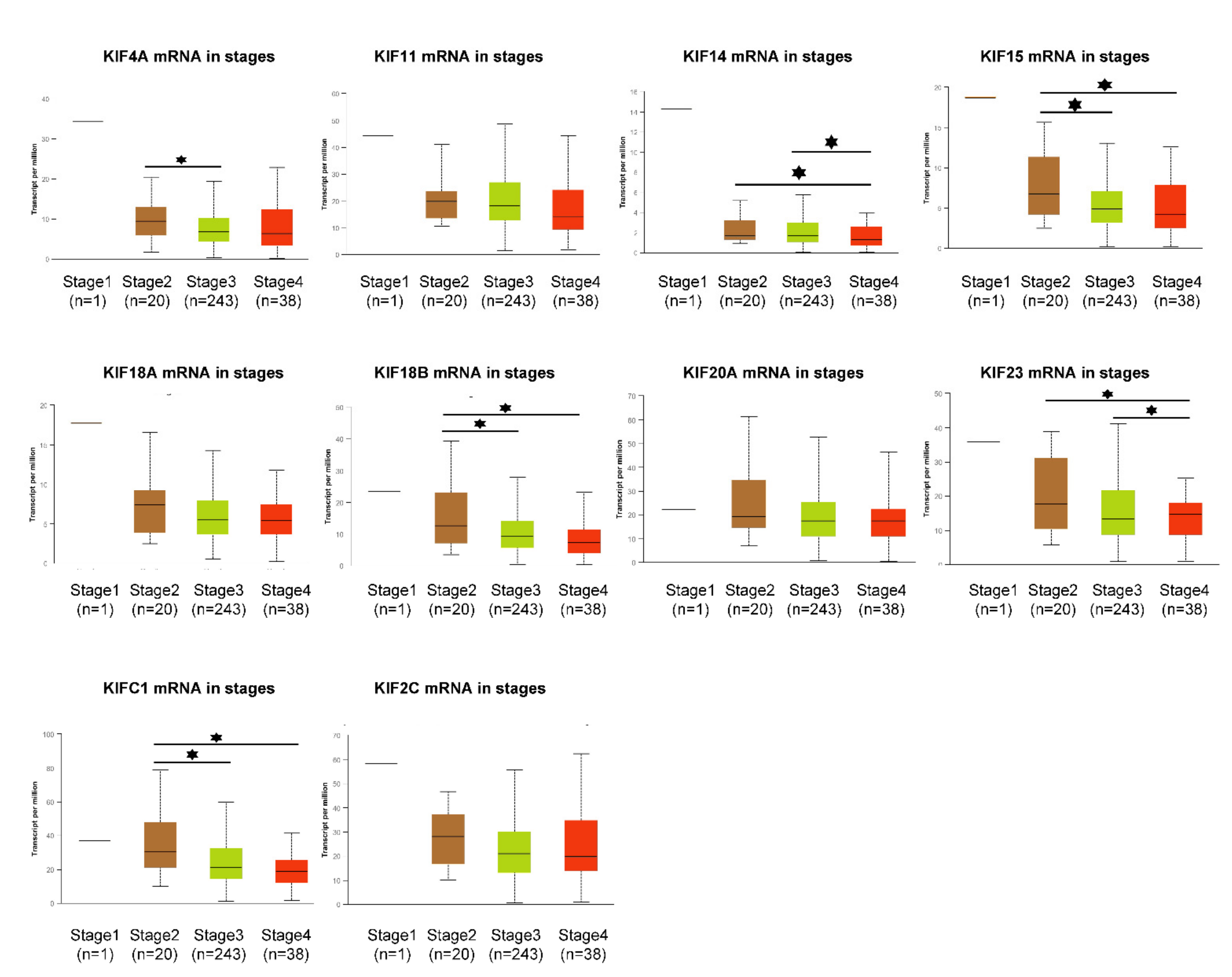
3.6. Expression Profile of Overexpressed Mitotic KIFs in Different OC Stages and Grades
3.7. Overexpression of Survival-Related Mitotic KIFs Indicates Worse Prognoses in Early-Stage and Low-Grade OC Patients
3.8. Overexpression of Survival-Related Mitotic KIFs May Related to Chemoresistance
4. Discussion
4.1. A New Insight Connects Mitotic KIFs with OC
4.2. Hypothesis I: Survival-Related Mitotic KIFs Are the Potential Prognostic Biomarkers for the OC
4.3. Hypothesis II: Several Mitotic KIFs May Be Promising Therapeutic Targets for the OC Treatment
4.4. Hypothesis III: Inherent Resistance of OC through Reduced Immonosurveillance and Subpopulations of Drug-Resistant OC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, H.; Takei, J. Management of hereditary breast and ovarian cancer. Int. J. Clin. Oncol. 2018, 23, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Nash, Z.; Menon, U. Ovarian cancer screening: Current status and future directions. Best Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Norouzi-Barough, L.; Sarookhani, M.R.; Sharifi, M.; Moghbelinejad, S.; Jangjoo, S.; Salehi, R. Molecular mechanisms of drug resistance in ovarian cancer. J. Cell Physiol. 2018, 233, 4546–4562. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Nag, S.; Aggarwal, S.; Rauthan, A.; Warrier, N. Maintenance therapy for recurrent epithelial ovarian cancer: Current therapies and future perspectives-a review. J. Ovarian Res. 2019, 12, 103. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Wang, X.; Liotta, L. Clinical bioinformatics: A new emerging science. J. Clin. Bioinform. 2011, 1, 1. [Google Scholar] [CrossRef] [Green Version]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef] [Green Version]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Rath, O.; Kozielski, F. Kinesins and cancer. Nat. Rev. Cancer 2012, 12, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yang, L.; Zhang, Z.; Yu, J.; Dai, L.; Gao, M.; Shang, Z.; Niu, Y. KIF20A Affects the Prognosis of Bladder Cancer by Promoting the Proliferation and Metastasis of Bladder Cancer Cells. Dis. Markers 2019, 2019, 4863182. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Chai, C.; Shen, T.; Li, X.; Ji, J.; Li, C.; Shang, Z.; Niu, Y. Aberrant KIF20A Expression Is Associated with Adverse Clinical Outcome and Promotes Tumor Progression in Prostate Cancer. Dis. Markers 2019, 2019, 4782730. [Google Scholar] [CrossRef]
- Zhang, Q.; Di, J.; Ji, Z.; Mi, A.; Li, Q.; Du, X.; Wang, A.; Wang, A.; Qin, C. KIF20A Predicts Poor Survival of Patients and Promotes Colorectal Cancer Tumor Progression through the JAK/STAT3 Signaling Pathway. Dis. Markers 2020, 2020, 2032679. [Google Scholar] [CrossRef]
- Tischer, J.; Gergely, F. Anti-mitotic therapies in cancer. J. Cell Biol. 2019, 218, 10–11. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlen, M.; Fagerberg, L.; Fagerberg, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Khodabakhshi, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Hu, F.F.; Xia, M.X.; Han, L.; Zhang, Q.; Guo, A.Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Gao, J.J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gorss, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Andrews, L.; Mutch, D.G. Hereditary Ovarian Cancer and Risk Reduction. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 31–48. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, H.; Zhang, C.; Hu, T.; Wu, J.; Lin, X.; Luo, D.; Wang, C.; Meng, L.; Xi, L.; et al. Gene co-expression network reveals shared modules predictive of stage and grade in serous ovarian cancers. Oncotarget 2017, 8, 42983–42996. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.Y.; Ozcelik, H.; Cheung, A.N.; Ngan, H.Y.; Khoo, U.S. Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 2002, 62, 4151–4156. [Google Scholar]
- Kawai, Y.; Shibata, K.; Shibata, J.; Suzuki, S.; Utsumi, F.; Niimi, K.; Sekiya, R.; Senga, T.; Kikkawa, F.; Kajiyama, H. Kif20a Expression as a Prognostic Indicator and Its Possible Involvement in the Proliferation of Ovarian Clear-Cell Carcinoma. Int. J. Gynecol. Cancer 2018, 28, 167. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.; Carville, S.; McKenna, F.G. Guideline Development, Diagnosis and management of rheumatoid arthritis in adults: Summary of updated NICE guidance. BMJ 2018, 362, k3015. [Google Scholar]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Mikula-Pietrasik, J.; Witucka, A.; Witucka, M.; Witucka, P.; Begier-Krasińska, B.; Niklas, A.; Tykarski, A.; Książek, K. Comprehensive review on how platinum- and taxane-based chemotherapy of ovarian cancer affects biology of normal cells. Cell Mol. Life Sci. 2019, 76, 681–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, L.C.; Lu, K.H.; Linette, G.P.; Cliby, W.A.; Kalli, K.R.; Gershenson, D.; Bast, R.C.; Stec, J.; Lartchouk, N.; Smith, D.I.; et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin. Cancer Res. 2005, 11, 2149–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zhang, W.; Sun, X.; Chen, J.; Li, Y.; Niu, C.; Xu, B.; Zhang, Y. Overexpression of kinesin family member 20A is associated with unfavorable clinical outcome and tumor progression in epithelial ovarian cancer. Cancer Manag. Res. 2018, 10, 3433–3450. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Wang, W.; Hong, B.; Jiang, X.; Sun, R.; Yan, Q.; Zhang, S.; Lu, M.; Wang, S.; Zhang, Z.; et al. Upregulation of KIF20A correlates with poor prognosis in gastric cancer. Cancer Manag. Res. 2018, 10, 6205–6216. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Takano, A.; Thang, P.M.; Tsevegjav, B.; Zhu, M.; Yokose, T.; Yokose, T.; Miyagi, Y.; Daigo, Y. Characterization of KIF20A as a prognostic biomarker and therapeutic target for different subtypes of breast cancer. Int. J. Oncol. 2020, 57, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; He, C.; Gao, S.; Yang, Z.; Li, L.; Qiao, L.; Fang, L. KIF20A silence inhibits the migration, invasion and proliferation of non-small cell lung cancer and regulates the JNK pathway (Retracted article. See vol. 48, pg. 157, 2021). Clin. Exp. Pharmacol. Physiol. 2020, 47, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.; Donthamsetty, S.; Pannu, V.; Rida, P.; Ogden, A.; Bowen, N.; Osan, R.; Cantuaria, G.; Aneja, R. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: Delineating protein interaction networks and signaling circuitries. J. Ovarian Res. 2014, 7, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Zhu, H.; Ye, Q.; Wang, C.; Xu, Y. Prognostic Value of KIF2A and HER2-Neu Overexpression in Patients With Epithelial Ovarian Cancer. Medicine (Baltimore) 2016, 95, e2803. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.L.; Deng, S.Z.; Li, C.; Tian, Z.N.; Song, X.Q.; Yao, G.D.; Geng, J.S. High expression of KIF14 is associated with poor prognosis in patients with epithelial ovarian cancer. Eur. Rev. Med. Pharmaco. Sci. 2017, 21, 239–245. [Google Scholar]
- Yang, X.; Zhang, L.; Xie, L. Upregulation of KIF26B, Cell Migration and Proliferation of Human Ovarian Cancer Cell Lines In Vitro, and Patient Outcomes from Human Bioinformatic Analysis. Med. Sci. Monit. 2018, 24, 3863–3872. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D.; Reese, T.S.; Sheetz, M.P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 1985, 42, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Miki, H.; Setou, M.; Hirokawa, N. All kinesin superfamily protein, KIF, genes in the mouse and human genome and transcripts. Mol. Biol. Cell 2002, 13, 184a. [Google Scholar]
- Lucanus, A.J.; Yip, G.W. Kinesin superfamily: Roles in breast cancer, patient prognosis and therapeutics. Oncogene 2018, 37, 833–838. [Google Scholar] [CrossRef]
- Li, T.F.; Zeng, H.J.; Shan, Z.; Ye, R.Y.; Cheang, T.Y.; Zhang, Y.J.; Lu, S.H.; Zhang, Q.; Shao, N.; Lin, Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020, 20, 123. [Google Scholar] [CrossRef] [Green Version]
- Neijt, J.P.; Engelholm, S.A.; Tuxen, M.K.; Engelholm, P.G.; Hansen, M.; Sessa, C.; de Swart, C.A.; Hirsch, F.R.; Lund, B.; van Houwelingen, H.C. Exploratory phase III study of paclitaxel and cisplatin versus paclitaxel and carboplatin in advanced ovarian cancer. J. Clin. Oncol. 2000, 18, 3084–3092. [Google Scholar] [CrossRef]
- Ozols, R.F.; Bundy, B.N.; Greer, B.E.; Fowler, J.M.; Clarke-Pearson, D.; Burger, R.A.; Mannel, R.S.; DeGeest, K.; Hartenbach, E.M.; Hartenbach, R.; et al. Phase III trial of carboplatin and paclitaxel compared with cisplatin and paclitaxel in patients with optimally resected stage III ovarian cancer: A Gynecologic Oncology Group study. J. Clin. Oncol. 2003, 21, 3194–3200. [Google Scholar] [CrossRef]
- Agarwal, R.; Kaye, S.B. Ovarian cancer: Strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 2003, 3, 502–516. [Google Scholar] [CrossRef]
- Christie, E.L.; Bowtell, D.D.L. Acquired chemotherapy resistance in ovarian cancer. Ann. Oncol. 2017, 28, viii13–viii15. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Ray, C.M.; Spira, J.B.; Trepel, J.B.; Carter, C.A.; Cottrill, H.M. A brief review of the management of platinum-resistant-platinum-refractory ovarian cancer. Med. Oncol. 2017, 34, 103. [Google Scholar] [CrossRef]
- Walczak, C.E.; Gayek, S.; Ohi, R. Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 2013, 29, 417–441. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shu, K.P.; Wang, Z.F.; Ding, D.G. Prognostic significance of KIF2A and KIF20A expression in human cancer A systematic review and meta-analysis. Medicine 2019, 98, e18040. [Google Scholar]
- Yap, T.A.; Omlin, A.; de Bono, J.S. Development of therapeutic combinations targeting major cancer signaling pathways. J. Clin. Oncol. 2013, 31, 1592–1605. [Google Scholar] [CrossRef] [PubMed]
- Blagden, S.P.; Molife, L.R.; Seebaran, A.; Payne, M.; Reid, A.H.M.; Seebaran, A.S.; Seebaran, L.S.; Williams, D.D.; Bowen, C.; Kathman, S.J.; et al. A phase I trial of ispinesib, a kinesin spindle protein inhibitor, with docetaxel in patients with advanced solid tumours. Br. J. Cancer 2008, 98, 894–899. [Google Scholar] [CrossRef] [Green Version]
- Basso, A.D.; Liu, M.; Dai, C.; Gray, K.; Nale, L.; Tevar, S.; Lee, S.; Liang, L.; Ponery, A.; Yaremko, B.; et al. SCH 2047069, a novel oral kinesin spindle protein inhibitor, shows single-agent antitumor activity and enhances the efficacy of chemotherapeutics. Mol. Cancer Ther. 2010, 9, 2993–3002. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.A.; Reader, J.; Roque, D.M. Review of Immune Therapies Targeting Ovarian Cancer. Curr. Treat. Options Oncol. 2018, 19, 74. [Google Scholar] [CrossRef]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin. Cancer Biol. 2019, 59, 147–160. [Google Scholar] [CrossRef]
- Zheng, F.; Zhang, Y.; Chen, S.; Weng, X.; Rao, Y.; Fang, H. Mechanism and current progress of Poly ADP-ribose polymerase (PARP) inhibitors in the treatment of ovarian cancer. Biomed. Pharm. 2020, 123, 109661. [Google Scholar] [CrossRef]
- Burotto, M.; Chiou, V.L.; Lee, J.M.; Kohn, E.C. The MAPK pathway across different malignancies: A new perspective. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, X.H.; Li, S.; Xu, B.Z.; Wang, Z.B.; Quan, S.; Li, M.; Zhang, Q.H.; Ouyang, Y.C.; Schatten, H.; Xing, F.Q.; et al. p38 alpha MAPK is a MTOC-associated protein regulating spindle assembly, spindle length and accurate chromosome segregation during mouse oocyte meiotic maturation. Cell Cycle 2010, 9, 4130–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.J.; Yang, W.X. Kinesins in MAPK cascade: How kinesin motors are involved in the MAPK pathway? Gene 2019, 684, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Patch, A.M.; Christie, E.L.; Bowtell, D.D.L. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Lagana, A.S.; Sofo, V.; Vitale, S.G.; Triolo, O. Epithelial ovarian cancer inherent resistance: May the pleiotropic interaction between reduced immunosurveillance and drug-resistant cells play a key role? Gynecol. Oncol. Rep. 2016, 18, 57–58. [Google Scholar] [CrossRef] [Green Version]
- Lagana, A.S.; Colonese, F.; Colonese, E.; Sofo, V.; Salmeri, F.M.; Granese, R.; Chiofalo, B.; Ciancimino, L.; Triolo, O. Cytogenetic analysis of epithelial ovarian cancer’s stem cells: An overview on new diagnostic and therapeutic perspectives. Eur. J. Gynaecol. Oncol. 2015, 36, 495–505. [Google Scholar]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial ovarian cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Annett, S.L.; Morgan, M.P.; Robson, T. The Cancer Stem Cell Niche in Ovarian Cancer and Its Impact on Immune Surveillance. Int. J. Mol. Sci. 2021, 22, 4091. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [Green Version]




| Characteristics | All Patients (N) | All Patients (%) |
|---|---|---|
| Age at diagnosis median, years | ||
| <58 | 263 | 45.1 |
| ≥58 | 321 | 54.9 |
| FIGO stage | ||
| Early (I–II) | 46 | 7.9 |
| Late (III–IV) | 535 | 91.6 |
| NA | 3 | 0.5 |
| Histologic grade | ||
| Low (G1–G2) | 77 | 13.2 |
| High (G3) | 505 | 86.5 |
| NA | 2 | 0.30 |
| OS status | ||
| Living | 227 | 38.9 |
| Deceased | 351 | 60.1 |
| NA | 6 | 1.0 |
| OS median, months | ||
| <32 | 285 | 48.8 |
| ≥32 | 296 | 50.7 |
| NA | 3 | 0.5 |
| PFS median, months | ||
| <14 | 239 | 41.0 |
| ≥14 | 259 | 44.3 |
| NA | 86 | 14.7 |
| Gene Symbol Description | Ensemble ID | Chromosome | Transcripts Number | Gene Type | Protein Function |
|---|---|---|---|---|---|
| BLM | ENSG00000197299 | 15 | 11 | Protein coding | DNA replication/repair, genome integrity |
| AURKB | ENSG00000178999 | 17 | 14 | Protein coding | Cell-cycle regulation |
| LRRC8E | ENSG00000171017 | 19 | 6 | Protein coding | Anion/aspartate transmembrane transport |
| METTL7B | ENSG00000170439 | 12 | 2 | Protein coding | Methyltransferase activity |
| GPRIN1 | ENSG00000169258 | 5 | 1 | Protein coding | Neurite outgrowth, phosphoprotein binding |
| BUB1B | ENSG00000156970 | 15 | 11 | Protein coding | Mitosis progression, ATP binding |
| BRCA2 | ENSG00000139618 | 13 | 11 | Protein coding | Double-strand break repair/homologous recombination |
| E2F8 | ENSG00000129173 | 11 | 5 | Protein coding | DNA binding transcription factor activity |
| MYCL | ENSG00000116990 | 1 | 4 | Protein coding | DNA binding, protein dimerization activity |
| KIF20A | ENSG00000112984 | 5 | 7 | Protein coding | Microtubule binding, ATPase activity |
| ICAM3 | ENSG00000076663 | 19 | 10 | Protein coding | Integrin and signaling receptor binding |
| GTSE1 | ENSG00000075218 | 22 | 4 | Protein coding | P53-induced cell-cycle arrest, Microtubule binding |
| HMMR | ENSG00000072571 | 5 | 8 | Protein coding | Cell motility, cellular transformation, metastasis formation |
| Mitotic KIFs | Kinesins Family | Localization | Cell Cycle Stage | Main Function |
|---|---|---|---|---|
| KIF11 | Kinesin-5 | Spindle/pole | Prophase, Metaphase | Bipolar spindle formation, separation of duplicated centrosomes |
| KIF14 (CMKRP) | Kinesin-3 | Spindle/midbody | Telophase, Cytokinesis | Cytokinesis, chromosome congression and alignment |
| KIF15 (HKLP2) | Kinesin-12 | Spindle/pole/midzone | Metaphase | Bipolar spindle formation in absence of KIF11 |
| KIF18B | Kinesin-8 | Spindle/pole | Interphase, Metaphase | Chromosome congression and alignment microtubule depolymerization |
| KIF20A (MKLP2, RabK6) | Kinesin-6 | Spindle/midzone /midbody | Anaphase, Cytokinesis | Cytokinesis |
| KIF23 (MKLP1, KNSL5) | Kinesin-6 | Spindle/midzone /midbody | Telophase, Cytokinesis | Cytokinesis |
| KIFC1 (HEST, KNSL2) | Kinesin-14 | Spindle/pole | Prophase | Chromosome congression and alignment bipolar spindle formation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Chen, C.; Fehm, T.; Cheng, Z.; Neubauer, H. Identifying Mitotic Kinesins as Potential Prognostic Biomarkers in Ovarian Cancer Using Bioinformatic Analyses. Diagnostics 2022, 12, 470. https://doi.org/10.3390/diagnostics12020470
Liu H, Chen C, Fehm T, Cheng Z, Neubauer H. Identifying Mitotic Kinesins as Potential Prognostic Biomarkers in Ovarian Cancer Using Bioinformatic Analyses. Diagnostics. 2022; 12(2):470. https://doi.org/10.3390/diagnostics12020470
Chicago/Turabian StyleLiu, Hailun, Chen Chen, Tanja Fehm, Zhongping Cheng, and Hans Neubauer. 2022. "Identifying Mitotic Kinesins as Potential Prognostic Biomarkers in Ovarian Cancer Using Bioinformatic Analyses" Diagnostics 12, no. 2: 470. https://doi.org/10.3390/diagnostics12020470






