Diagnostic Performance of Selected MRI-Derived Radiomics Able to Discriminate Progression-Free and Overall Survival in Patients with Midline Glioma and the H3F3AK27M Mutation
Abstract
:1. Introduction
2. Materials and Methods
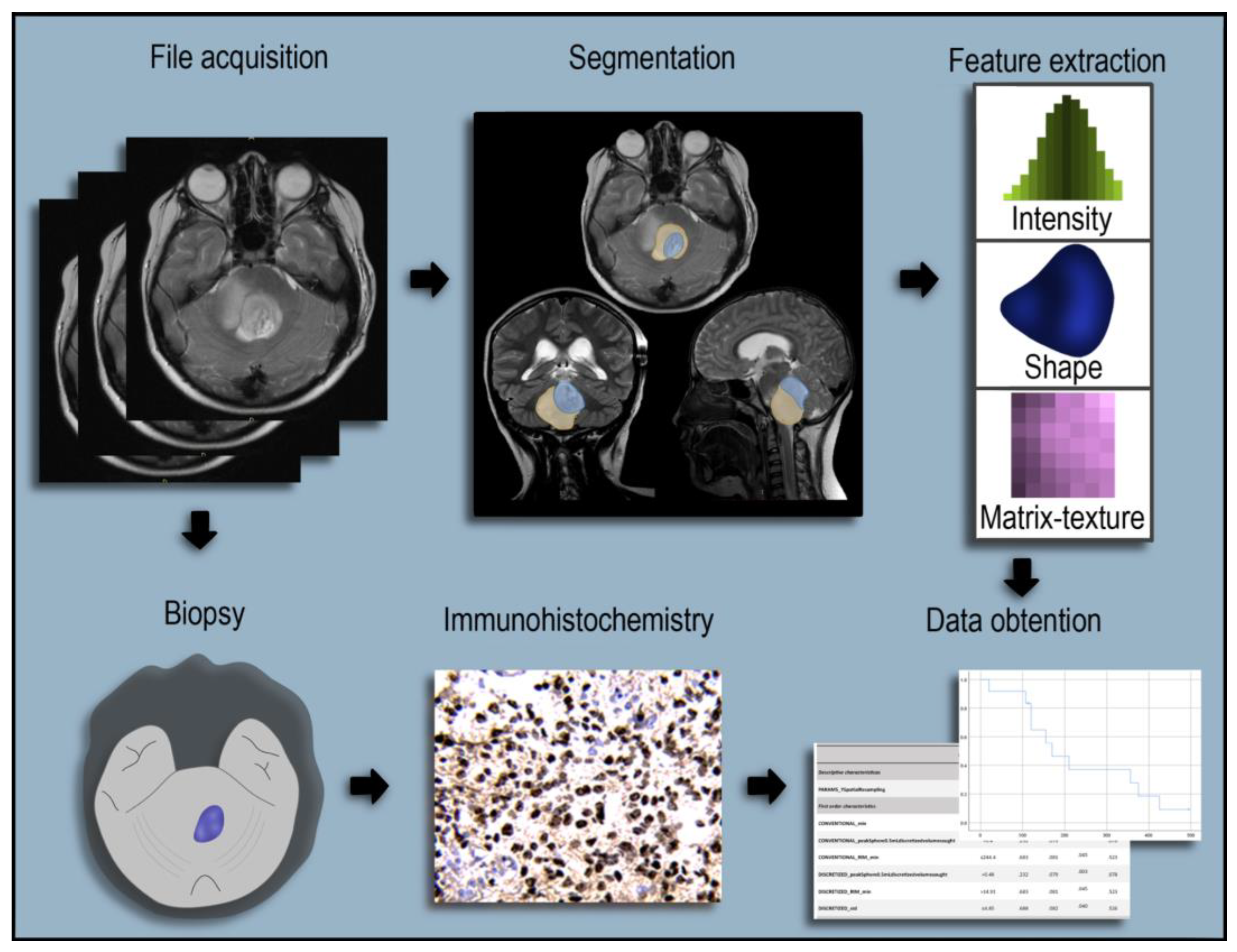
2.1. Imaging Evaluation of Midline Gliomas
2.2. Radiomics Extraction
2.3. Definitions of Progression-Free Survival and Overall Survival
2.4. Statistical Analyses
2.4.1. Descriptive Statistics and Differences between Tumour Regions
2.4.2. Evaluation of Diagnostic Performance
2.4.3. Software
3. Results
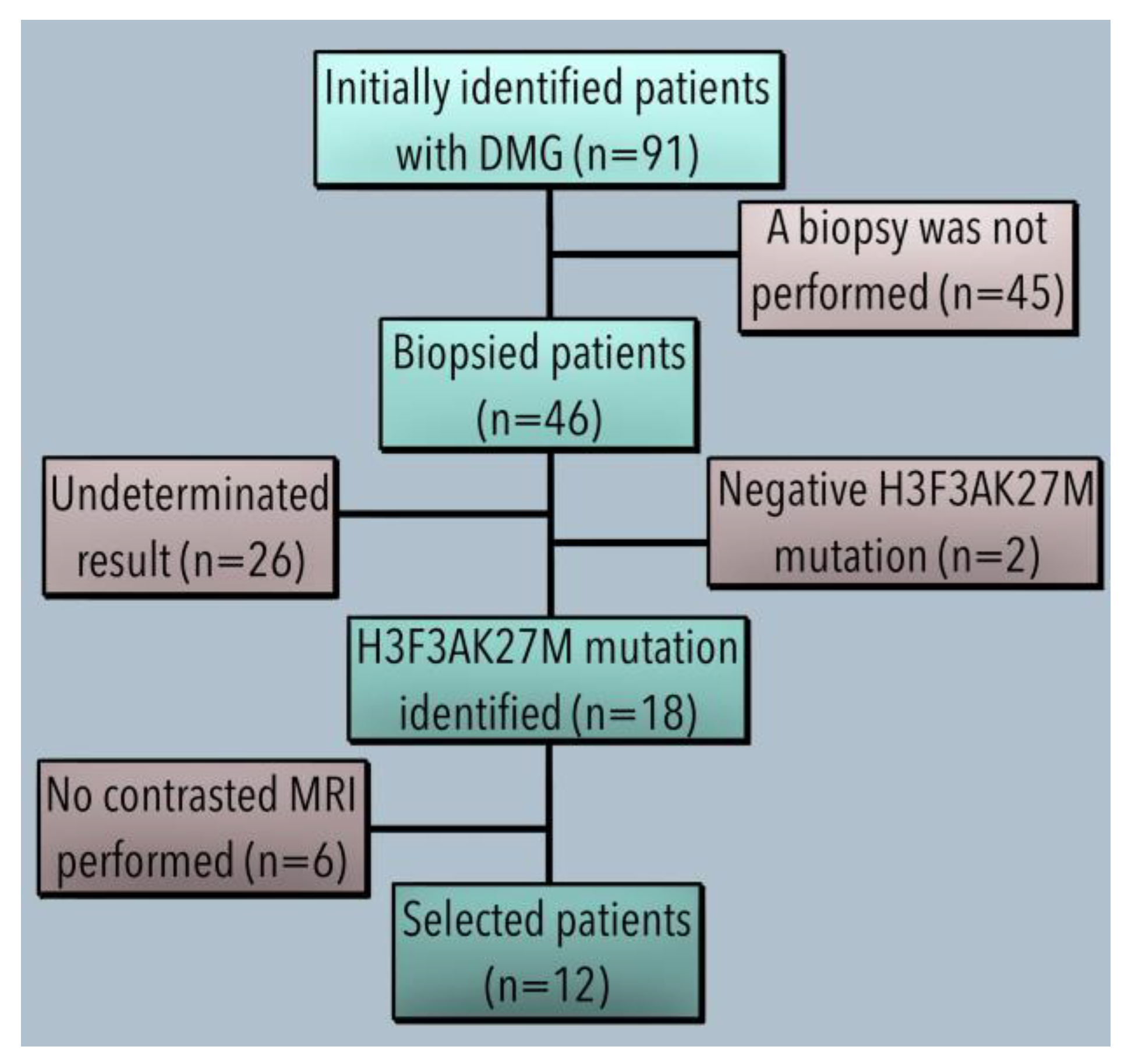
3.1. Demographics and Clinical Features
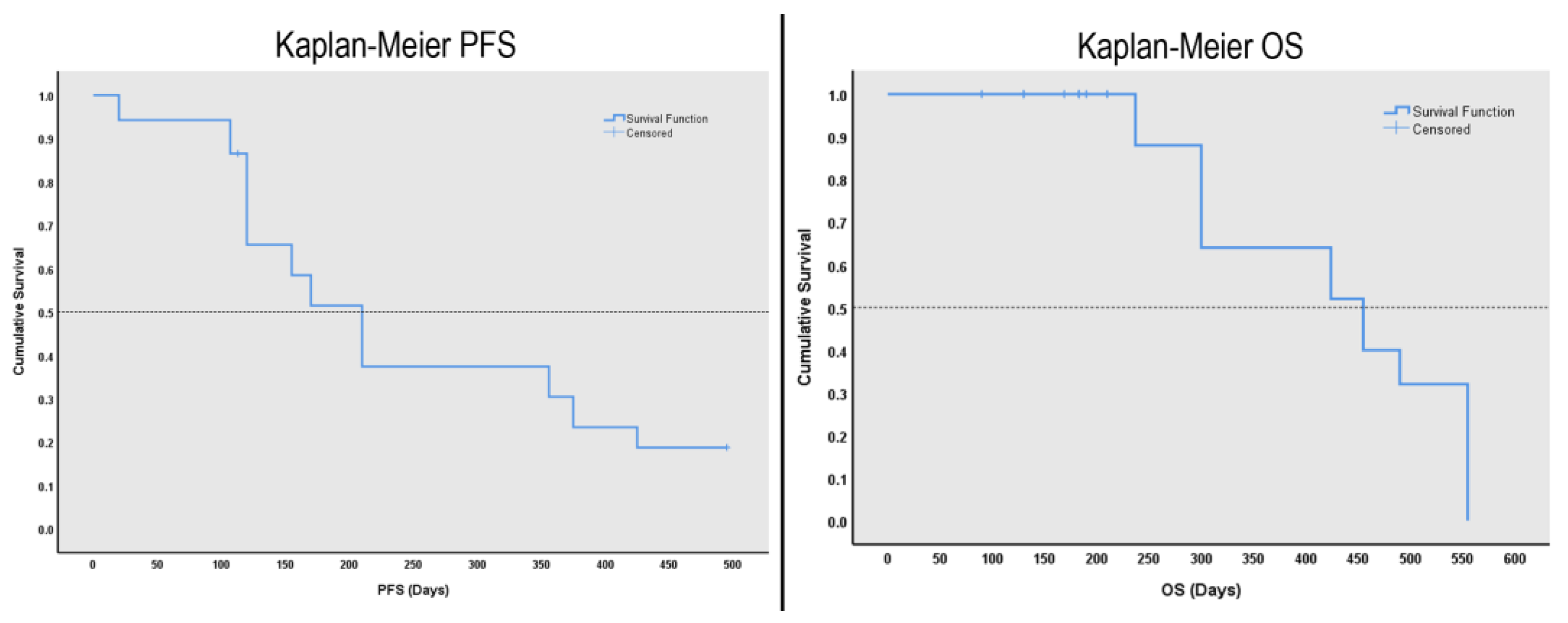
3.2. Survival Analysis
3.3. Radiomic Values Differences between Tumour Regions
3.4. Radiomic Values Extraction
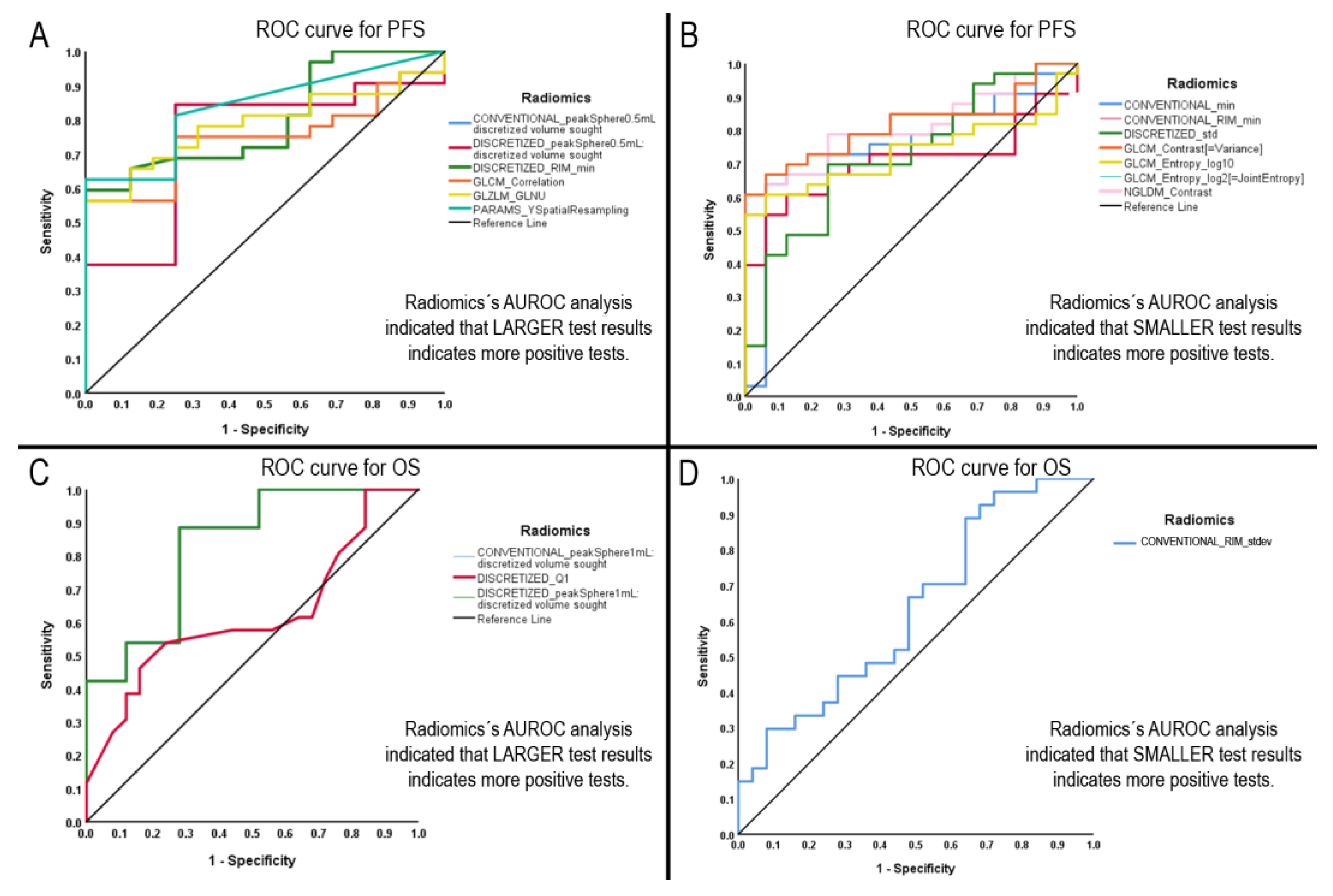
3.5. Radiomics with Significant AUROCs to Identify PFS and OS above the Median
3.6. Diagnostic Performance Test
4. Discussion
4.1. Limitations of the Study
4.2. Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Ruscio, V.; Del Baldo, G.; Fabozzi, F.; Vinci, M.; Cacchione, A.; de Billy, E.; Megaro, G.; Carai, A.; Mastronuzzi, A. Pediatric Diffuse Midline Gliomas: An Unfinished Puzzle. Diagnostics 2022, 12, 2064. [Google Scholar] [CrossRef] [PubMed]
- Gianno, F.; Giovannoni, I.; Cafferata, B.; Diomedi-Camassei, F.; Minasi, S.; Barresi, S.; Buttarelli, F.R.; Alesi, V.; Cardoni, A.; Antonelli, M.; et al. Paediatric-type diffuse high-grade gliomas in the 5th CNS WHO Classification. Pathologica 2022, 114, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e5. [Google Scholar] [CrossRef] [Green Version]
- Coleman, C.; Chen, K.; Lu, A.; Seashore, E.; Stoller, S.; Davis, T.; Braunstein, S.; Gupta, N.; Mueller, S. Interdisciplinary care of children with diffuse midline glioma. Neoplasia 2023, 35, 100851. [Google Scholar] [CrossRef]
- Li, Q.; Dong, F.; Jiang, B.; Zhang, M. Exploring MRI Characteristics of Brain Diffuse Midline Gliomas With the H3 K27M Mutation Using Radiomics. Front. Oncol. 2021, 11, 646267. [Google Scholar] [CrossRef]
- Singh, G.; Manjila, S.; Sakla, N.; True, A.; Wardeh, A.H.; Beig, N.; Vaysberg, A.; Matthews, J.; Prasanna, P.; Spektor, V. Radiomics and radiogenomics in gliomas: A contemporary update. Br. J. Cancer 2021, 125, 641–657. [Google Scholar] [CrossRef]
- Preuss, K.; Thach, N.; Liang, X.; Baine, M.; Chen, J.; Zhang, C.; Du, H.; Yu, H.; Lin, C.; Hollingsworth, M.A.; et al. Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications. Cancers 2022, 14, 1654. [Google Scholar] [CrossRef]
- Miranda, J.; Horvat, N.; Fonseca, G.M.; Araujo-Filho, J.D.A.B.; Fernandes, M.C.; Charbel, C.; Chakraborty, J.; Coelho, F.F.; Nomura, C.H.; Herman, P. Current status and future perspectives of radiomics in hepatocellular carcinoma. World J. Gastroenterol. 2023, 29, 43–60. [Google Scholar] [CrossRef]
- Granata, V.; Caruso, D.; Grassi, R.; Cappabianca, S.; Reginelli, A.; Rizzati, R.; Masselli, G.; Golfieri, R.; Rengo, M.; Regge, D.; et al. Structured Reporting of Rectal Cancer Staging and Restaging: A Consensus Proposal. Cancers 2021, 13, 2135. [Google Scholar] [CrossRef]
- Ma, Y.; Lin, Y.; Lu, J.; He, Y.; Shi, Q.; Liu, H.; Li, J.; Zhang, B.; Zhang, J.; Zhang, Y.; et al. A meta-analysis of based radiomics for predicting lymph node metastasis in patients with biliary tract cancers. Front. Surg. 2022, 9, 1045295. [Google Scholar] [CrossRef]
- Gu, J.; Jiang, T. Ultrasound radiomics in personalised breast management: Current status and future prospects. Front. Oncol. 2022, 12, 963612. [Google Scholar] [CrossRef]
- Coppola, F.; Faggioni, L.; Gabelloni, M.; De Vietro, F.; Mendola, V.; Cattabriga, A.; Cocozza, M.A.; Vara, G.; Piccinino, A.; Monaco, S.L.; et al. Human, All Too Human? An All-Around Appraisal of the “Artificial Intelligence Revolution” in Medical Imaging. Front. Psychol. 2021, 12, 710982. [Google Scholar] [CrossRef]
- Kesler, S.R.; Harrison, R.A.; Rao, V.; Dyson, H.; Petersen, M.; Prinsloo, S. Predicting overall survival in diffuse glioma from the presurgical connectome. Sci. Rep. 2022, 12, 18783. [Google Scholar] [CrossRef]
- Su, X.; Chen, N.; Sun, H.; Liu, Y.; Yang, X.; Wang, W.; Zhang, S.; Tan, Q.; Su, J.; Gong, Q.; et al. Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro Oncol. 2020, 22, 393–401. [Google Scholar] [CrossRef]
- Kandemirli, S.G.; Kocak, B.; Naganawa, S.; Ozturk, K.; Yip, S.S.; Chopra, S.; Rivetti, L.; Aldine, A.S.; Jones, K.; Cayci, Z.; et al. Machine Learning-Based Multiparametric Magnetic Resonance Imaging Radiomics for Prediction of H3K27M Mutation in Midline Gliomas. World Neurosurg. 2021, 151, e78–e85. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Wen, L.-H.; Wu, G.; Hu, M.-Z.; Zhang, C.-C.; Chen, F.; Zhao, J.-N. Comparison of Radiomics Analyses Based on Different Magnetic Resonance Imaging Sequences in Grading and Molecular Genomic Typing of Glioma. J. Comput. Assist. Tomogr. 2021, 45, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, Q.; Wen, J.; Li, M.; Zhu, Y.; Liu, Y.; Dai, Y.; Guan, Y.; Zhou, Z.; Hua, T. Integrated CT Radiomics Features Could Enhance the Efficacy of (18)F-FET PET for Non-Invasive Isocitrate Dehydrogenase Genotype Prediction in Adult Untreated Gliomas: A Retrospective Cohort Study. Front. Oncol. 2021, 11, 772703. [Google Scholar] [CrossRef] [PubMed]
- Tam, L.T.; Yeom, K.W.; Wright, J.N.; Jaju, A.; Radmanesh, A.; Han, M.; Toescu, S.; Maleki, M.; Chen, E.; Campion, A.; et al. MRI-based radiomics for prognosis of pediatric diffuse intrinsic pontine glioma: An international study. Neurooncol. Adv. 2021, 3, vdab042. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zheng, H.; Li, J.; Zhang, Y.; Duan, S.; Li, Y.; Wang, D. MRI-based radiomics signature and clinical factor for predicting H3K27M mutation in pediatric high-grade gliomas located in the midline of the brain. Eur. Radiol. 2022, 32, 1813–1822. [Google Scholar] [CrossRef]
- Guo, W.; She, D.; Xing, Z.; Lin, X.; Wang, F.; Song, Y.; Cao, D. Multiparametric MRI-Based Radiomics Model for Predicting H3 K27M Mutant Status in Diffuse Midline Glioma: A Comparative Study Across Different Sequences and Machine Learning Techniques. Front. Oncol. 2022, 12, 796583. [Google Scholar] [CrossRef]
- Deng, D.-B.; Liao, Y.-T.; Zhou, J.-F.; Cheng, L.-N.; He, P.; Wu, S.-N.; Wang, W.-S.; Zhou, Q. Non-Invasive Prediction of Survival Time of Midline Glioma Patients Using Machine Learning on Multiparametric MRI Radiomics Features. Front. Neurol. 2022, 13, 866274. [Google Scholar] [CrossRef]
- Wagner, M.W.; Namdar, K.; Napoleone, M.; Hainc, N.; Amirabadi, A.; Fonseca, A.; Laughlin, S.; Shroff, M.M.; Bouffet, E.; Hawkins, C.; et al. Radiomic Features Based on MRI Predict Progression-Free Survival in Pediatric Diffuse Midline Glioma/Diffuse Intrinsic Pontine Glioma. Can. Assoc. Radiol. J. 2023, 74, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterisation of Tumor Heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef] [Green Version]
- Nioche, C. LIFExAnalytics 2023. Available online: https://www.lifexsoft.org/ (accessed on 27 January 2023).
- Durand-Munoz, C.; Flores-Alvarez, E.; Moreno-Jimenez, S.; Roldan-Valadez, E. Pre-operative apparent diffusion coefficient values and tumour region volumes as prognostic biomarkers in glioblastoma: Correlation and progression-free survival analyses. Insights Imaging 2019, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Flores-Alvarez, E.; Durand-Muñoz, C.; Cortes-Hernandez, F.; Muñoz-Hernandez, O.; Moreno-Jimenez, S.; Roldan-Valadez, E. Clinical Significance of Fractional Anisotropy Measured in Peritumoral Edema as a Biomarker of Overall Survival in Glioblastoma: Evidence Using Correspondence Analysis. Neurol. India 2019, 67, 1074–1081. [Google Scholar]
- Cortez-Conradis, D.; Favila, R.; Isaac-Olive, K.; Martinez-Lopez, M.; Rios, C.; Roldan-Valadez, E. Diagnostic performance of regional DTI-derived tensor metrics in glioblastoma multiforme: Simultaneous evaluation of p, q, L, Cl, Cp, Cs, RA, RD, AD, mean diffusivity and fractional anisotropy. Eur. Radiol. 2013, 23, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Archila-Rincon, L.M.; Del Carmen Garcia-Blanco, M.; Roldan-Valadez, E. Diagnostic performance of CT densities in selected gray- and white-matter regions for the clinical diagnosis of brain death: A retrospective study in a tertiary-level general hospital. Eur. J. Radiol. 2018, 108, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Goo, J.M.; Jo, C.-H. Receiver operating characteristic (ROC) curve: Practical review for radiologists. Korean J. Radiol. 2004, 5, 11–18. [Google Scholar] [CrossRef] [Green Version]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Metz, C.E. Basic principles of ROC analysis. Semin. Nucl. Med. 1978, 8, 283–298. [Google Scholar] [CrossRef]
- Xie, T.; Chen, X.; Fang, J.; Kang, H.; Xue, W.; Tong, H.; Cao, P.; Wang, S.; Yang, Y.; Zhang, W. Textural features of dynamic contrast-enhanced MRI derived model-free and model-based parameter maps in glioma grading. J. Magn. Reson. Imaging 2018, 47, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.X.; Shi, D.F.; Ren, S.X.; Zhang, S.Y.; Li, L.; Li, Q.C.; Guan, L.M. Histogram analysis of diffusion kurtosis imaging derived maps may distinguish between low and high grade gliomas before surgery. Eur. Radiol. 2018, 28, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.B.; Choi, Y.S.; Bae, S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur. Radiol. 2018, 28, 3832–3839. [Google Scholar] [CrossRef]
- Taylor, C.; Ekert, J.O.; Sefcikova, V.; Fersht, N.; Samandouras, G. Discriminators of pseudoprogression and true progression in high-grade gliomas: A systematic review and meta-analysis. Sci. Rep. 2022, 12, 13258. [Google Scholar] [CrossRef]
- Molina, D.; Pérez-Beteta, J.; Luque, B.; Arregui, E.; Calvo, M.; Borrás, J.M.; López, C.; Martino, J.; Velasquez, C.; Asenjo, B.; et al. Tumour heterogeneity in glioblastoma assessed by MRI texture analysis: A potential marker of survival. Br. J. Radiol. 2016, 89, 20160242. [Google Scholar] [CrossRef] [Green Version]
- Shur, J.D.; Doran, S.J.; Kumar, S.; ap Dafydd, D.; Downey, K.; O’Connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. Radiographics 2021, 41, 1717–1732. [Google Scholar] [CrossRef]
- Soni, N.; Priya, S.; Bathla, G. Texture Analysis in Cerebral Gliomas: A Review of the Literature. AJNR Am. J. Neuroradiol. 2019, 40, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Tomaszewski, M.R.; Gillies, R.J. The Biological Meaning of Radiomic Features. Radiology 2021, 299, E256. [Google Scholar] [CrossRef]
- Lohmann, P.; Franceschi, E.; Vollmuth, P.; Dhermain, F.; Weller, M.; Preusser, M.; Smits, M.; Galldiks, N. Radiomics in neuro-oncological clinical trials. Lancet Digit. Health 2022, 4, e841–e849. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- O’Byrne, M.; Ghosh, B.; Pakrashi, V.; Schoefs, F. Texture Analysis based Detection and Classification of Surface Features on Ageing Infrastructure Elements. In BCRI2012 Bridge & Concrete Research in Ireland; HAL Open Science: Lyon, France, 2012. [Google Scholar]
- Syed, A.; Adam, R.; Ren, T.; Lu, J.; Maldjian, T.; Duong, T.Q. Machine learning with textural analysis of longitudinal multiparametric MRI and molecular subtypes accurately predicts pathologic complete response in patients with invasive breast cancer. PLoS ONE 2023, 18, e0280320. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Folty, M.; et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: A multicentre, retrospective study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B.; Zhang, Z.; Shi, X.; Cao, C.; Zhang, Z.; Hu, Z.; Ji, N.; Tian, J. Real-time intraoperative glioma diagnosis using fluorescence imaging and deep convolutional neural networks. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 3482–3492. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, Y.; Yang, J.J.; Zhao, J.; Xiang, Y.; Xia, C.; Cao, Y.; Chen, B.; Guan, H.; Qi, Y.-F.; et al. MRI-based radiomics analysis improves pre-operative diagnostic performance for the depth of stromal invasion in patients with early stage cervical cancer. Insights Imaging 2022, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Shiradkar, R.; Ghose, S.; Jambor, I.; Taimen, P.; Ettala, O.; Purysko, A.S.; Madabhushi, A. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: Preliminary findings. J. Magn. Reson. Imaging 2018, 48, 1626–1636. [Google Scholar] [CrossRef]
- Moradmand, H.; Aghamiri, S.M.R.; Ghaderi, R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020, 21, 179–190. [Google Scholar] [CrossRef]
- Um, H.; Tixier, F.; Bermudez, D.; Deasy, J.O.; Young, R.J.; Veeraraghavan, H. Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys. Med. Biol. 2019, 64, 165011. [Google Scholar] [CrossRef]
- Waugh, S.A.; Lerski, R.A.; Bidaut, L.; Thompson, A.M. The influence of field strength and different clinical breast MRI protocols on the outcome of texture analysis using foam phantoms. Med. Phys. 2011, 38, 5058–5066. [Google Scholar] [CrossRef] [Green Version]
- Buch, K.; Kuno, H.; Qureshi, M.M.; Li, B.; Sakai, O. Quantitative variations in texture analysis features dependent on MRI scanning parameters: A phantom model. J. Appl. Clin. Med. Phys. 2018, 19, 253–264. [Google Scholar] [CrossRef] [Green Version]
- Saint Martin, M.J.; Orlhac, F.; Akl, P.; Khalid, F.; Nioche, C.; Buvat, I.; Malhaire, C.; Frouin, F. A radiomics pipeline dedicated to Breast MRI: Validation on a multi-scanner phantom study. MAGMA 2021, 34, 355–366. [Google Scholar] [CrossRef]
- Shinohara, R.T.; Sweeney, E.M.; Goldsmith, J.; Shiee, N.; Mateen, F.J.; Calabresi, P.A.; Jarso, S.; Pham, D.L.; Reich, D.S.; Crainiceanu, C.M.; et al. Corrigendum to “Statistical normalisation techniques for magnetic resonance imaging” [NeuroImage: Clinical 6 (2014) 9-19]. Neuroimage Clin. 2015, 7, 848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.; Chan, S.; Chen, J.-H.; Chang, D.; Nie, K.; Chen, S.-T.; Lin, C.-J.; Shih, T.-C.; Nalcioglu, O.; Su, M.-Y. A new bias field correction method combining N3 and FCM for improved segmentation of breast density on MRI. Med. Phys. 2011, 38, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.P.; Sweeney, E.M.; Muschelli, J.; Crainiceanu, C.M.; Shinohara, R.T.; Alzheimer’s Disease Neuroimaging Initiative. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage 2016, 132, 198–212. [Google Scholar] [CrossRef] [Green Version]

| Radiomic Features | Percentile | |||
|---|---|---|---|---|
| Median | 75 | 25 | IQR | |
| CONVENTIONAL_min | 128.294 | 189.296 | 4.032 | 185.264 |
| CONVENTIONAL_peakSphere0.5mL: discretised volume sought | 0.506 | 0.522 | 0.472 | 0.05 |
| CONVENTIONAL_RIM_min | 257.726 | 337.008 | 160.108 | 176.972 |
| CONVENTIONAL_RIM_stdev | 70.049 | 106.502 | 32.110 | 74.392 |
| CONVENTIONAL_RIM_Volume(vx) | 1528.486 | 2413.208 | 783.969 | 1629.293 |
| DISCRETIZED_std | 4.516 | 5.733 | 2.995 | 2.738 |
| DISCRETIZED_Q1 | 15.500 | 19.75 | 10 | 9.75 |
| DISCRETIZED_peakSphere0.5mL: discretised volume sought | 0.506 | 0.522 | 0.472 | 0.05 |
| DISCRETIZED_peakSphere1mL: discretised volume sought | 1.041 | 1.045 | 1.014 | 0.031 |
| DISCRETIZED_RIM_min | 13.551 | 250.895 | 7.077 | 243.818 |
| PARAMS_YSpatialResampling | 0.654 | 4.864 | 0.5 | 4.364 |
| GLCM_Contrast[=Variance] | 10.566 | 22.434 | 0.046 | 22.388 |
| GLCM_Correlation | 0.606 | 5.745 | 0.484 | 5.261 |
| GLCM_Entropy_log10 | 2.071 | 2.368 | 0.567 | 1.801 |
| GLCM_Entropy_log2[=JointEntropy] | 6.878 | 7.868 | 2.230 | 5.638 |
| NGLDM_Contrast | 0.027 | 0.084 | 0.001 | 0.083 |
| GLZLM_GLNU | 169.238 | 68,327.15 | 65.305 | 68,261.845 |
| Radiomic Feature | Cut-Off Value | AUROC | SE | p-Value | 95% CI | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Progression-free survival | ||||||
| Descriptive characteristics | ||||||
| PARAMS_YSpatialResampling | >0.65 | 0.188 | 0.064 | 0.001 | 0.062 | 0.313 |
| First-order characteristics | ||||||
| CONVENTIONAL_min | ≤100.8 | 0.763 | 0.078 | 0.004 | 0.611 | 0.916 |
| CONVENTIONAL_peakSphere0.5mLdiscretizedvolumesought | >0.4 | 0.232 | 0.079 | 0.003 | 0.078 | 0.386 |
| CONVENTIONAL_RIM_min | ≤244.4 | 0.683 | 0.081 | 0.045 | 0.523 | 0.843 |
| DISCRETIZED_peakSphere0.5mLdiscretizedvolumesought | >0.48 | 0.232 | 0.079 | 0.003 | 0.078 | 0.386 |
| DISCRETIZED_RIM_min | >14.91 | 0.683 | 0.081 | 0.045 | 0.523 | 0.843 |
| DISCRETIZED_std | ≤4.85 | 0.688 | 0.082 | 0.040 | 0.526 | 0.849 |
| Second-order characteristics | ||||||
| GLCM_ContrastVariance | ≤6.12 | 0.781 | 0.069 | 0.002 | 0.647 | 0.916 |
| GLCM_Correlation | >0.69 | 0.290 | 0.077 | 0.022 | 0.138 | 0.442 |
| GLCM_Entropy_log10 | ≤1.57 | 0.685 | 0.079 | 0.043 | 0.530 | 0.841 |
| GLCM_Entropy_log2JointEntropy | ≤5.23 | 0.685 | 0.079 | 0.043 | 0.530 | 0.841 |
| GLZLM_GLNU | >232.13 | 0.246 | 0.072 | 0.005 | 0.104 | 0.387 |
| NGLDM_Contrast | ≤0.02 | 0.775 | 0.069 | 0.003 | 0.639 | 0.910 |
| Overall survival | ||||||
| First-order characteristics | ||||||
| CONVENTIONAL_ peakSphere1mLdiscretizedvolumesought | >1.02 | 0.134 | 0.054 | <0.001 | 0.029 | 0.240 |
| CONVENTIONAL_RIM_stdev | ≤106.56 | 0.686 | 0.080 | 0.035 | 0.529 | 0.843 |
| DISCRETIZED_Q1 | >17 | 0.321 | 0.083 | 0.042 | 0.160 | 0.483 |
| DISCRETIZED_ peakSphere1mLdiscretizedvolumesought | >1.02 | 0.134 | 0.054 | <0.001 | 0.029 | 0.240 |
| Radiomic (Significant AUROC) | Sensitivity | Specificity | +LR | −LR | +PV | −PV | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | 95% CI | Value | 95% CI | Value | 95% CI | Value | 95% CI | Value | 95% CI | Value | 95% CI | ||
| PFS | CONVENTIONAL_min | 63.8% | 46.2–79.1% | 93.7% | 69.7–99.8% | 10.2% | 1.5–69.2% | 0.3% | 0.2–0.6% | 95.8% | 77.2–99.3% | 53.7% | 42.3–64.4% |
| CONVENTIONAL_peakSphere0.5mLdiscretizedvolumesought | 97.2% | 85.4–99.9% | 0% | 0–20.5% | 0.9% | 0.9–1% | - | - | 68.6% | 67.4–69.8% | 0% | - | |
| CONVENTIONAL_RIM_min | 58.3% | 40.7–74.4% | 93.7% | 69.7–99.8% | 9.3% | 1.3–63.5% | 0.4% | 0.2–0.6% | 95.4% | 75.5–99.3% | 50% | 39.9–60% | |
| DISCRETIZED_peakSphere0.5mLdiscretizedvolumesought | 83.3% | 67.1–93.6% | 50% | 24.6–75.3% | 1.6% | 1–2.7% | 0.3% | 0.1–0.8% | 78.9% | 69.2–86.2% | 57.1% | 35.6–76.2% | |
| DISCRETIZED_ RIM_min | 63.8% | 46.2–79.1% | 93.7% | 69.7–99.8% | 10.2% | 1.5–69.2% | 0.3% | 0.2–0.6% | 95.8% | 77.2–99.3% | 53.5% | 42.3–64.4% | |
| DISCRETIZED_std | 63.8% | 46.2–79.1% | 75% | 47.6–92.7% | 2.5% | 1–6.1% | 0.4% | 0.2–0.8% | 85.1% | 70.3–93.2% | 48% | 35.4–60.7% | |
| GLCM_ ContrastVariance | 52.7% | 35.4–69.5% | 100% | 79.4–100% | - | - | 0.4% | 0.3–0.6% | 100% | - | 48.4% | 39.9–57.0% | |
| GLCM_Correlation | 52.7% | 35.4–69.5% | 93.7% | 69.7–99.8% | 8.4% | 1.2–57.7% | 0.5% | 0.3–0.7% | 95% | 73.5–99.2% | 46.8% | 37.9–56% | |
| GLCM_Entropy_log10 | 47.2% | 30.4–64.5% | 100% | 79.4–100% | - | - | 0.5% | 0.3–0.7% | 100% | - | 45.7% | 38.2–53.4% | |
| GLCM_Entropy_ log2JointEntropy | 47.2% | 30.4–64.5% | 100% | 79.4–100% | - | - | 0.5% | 0.3–0.7% | 100% | - | 45.7% | 38.2–53.4% | |
| GLZLM_GLNU | 52.7% | 35.4–69.5% | 93.7% | 69.7–99.8% | 8.4% | 1.2–57.7% | 0.5% | 0.3–0.7% | 95% | 73.5–99.2% | 46.8% | 37.9–56% | |
| NGLDM_Contrast | 55.5% | 38–72% | 93.7% | 69.7–99.8% | 8.8% | 1.3–60.6% | 0.4% | 0.3–0.6% | 95.2% | 74.5–99.2% | 48.3% | 38.9–57.9% | |
| PARAMS_ YSpatialResampling | 58.3% | 40.7–74.4% | 75% | 47.6–92.7% | 2.3% | 0.9–5.6% | 0.5% | 0.3–0.8% | 84% | 68.2–92.7% | 44% | 33.1–56.3% | |
| OS | CONVENTIONAL_ peakSphere1mLdiscretizedvolumesought | 85.1% | 66.6–95.8% | 60% | 38.6–78.8% | 2.1% | 1.2–3.5% | 0.2% | 0.09–0.6% | 69.6% | 58.1–79.2% | 78.9% | 58.9–90.7% |
| CONVENTIONAL_RIM_stdev | 85.1% | 66.2–95.8% | 36% | 17.9–57.4% | 1.3% | 0.9–1.8% | 0.4% | 0.1–1.1% | 58.9% | 50.7–66.7% | 69.2% | 44.1–86.4% | |
| DISCRETIZED_Q1 | 51.8% | 31.9–71.3% | 76% | 54.8–90.6% | 2.1% | 0.9–4.7% | 0.6% | 0.4–0.9% | 70% | 51.5–83.6% | 59.3% | 48.2–69.6% | |
| DISCRETIZED_ peakSphere1mLdiscretizedvolumesought | 85.1% | 66.2–95.8% | 60% | 38.6–78.8% | 2.1% | 1.2–3.5% | 0.2% | 0.09–0.6% | 69.6% | 58.1–79.2% | 78.9% | 58.9–84.4% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chilaca-Rosas, M.-F.; Garcia-Lezama, M.; Moreno-Jimenez, S.; Roldan-Valadez, E. Diagnostic Performance of Selected MRI-Derived Radiomics Able to Discriminate Progression-Free and Overall Survival in Patients with Midline Glioma and the H3F3AK27M Mutation. Diagnostics 2023, 13, 849. https://doi.org/10.3390/diagnostics13050849
Chilaca-Rosas M-F, Garcia-Lezama M, Moreno-Jimenez S, Roldan-Valadez E. Diagnostic Performance of Selected MRI-Derived Radiomics Able to Discriminate Progression-Free and Overall Survival in Patients with Midline Glioma and the H3F3AK27M Mutation. Diagnostics. 2023; 13(5):849. https://doi.org/10.3390/diagnostics13050849
Chicago/Turabian StyleChilaca-Rosas, Maria-Fatima, Melissa Garcia-Lezama, Sergio Moreno-Jimenez, and Ernesto Roldan-Valadez. 2023. "Diagnostic Performance of Selected MRI-Derived Radiomics Able to Discriminate Progression-Free and Overall Survival in Patients with Midline Glioma and the H3F3AK27M Mutation" Diagnostics 13, no. 5: 849. https://doi.org/10.3390/diagnostics13050849






