Comprehensive Analysis of Prognosis and Immune Landscapes Based on Lipid-Metabolism- and Ferroptosis-Associated Signature in Uterine Corpus Endometrial Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Information
2.2. Extraction of Lipid-Metabolism-Related and Ferroptosis-Associated Genes
2.3. Construction of the LMRG and FAR Prognostic Signature
2.4. Functional Enrichment Analysis
2.5. Tumor Mutational Burden (TMB) Analysis
2.6. Immune Infiltration of the Prognostic Model
2.7. Cell Culture
2.8. Small Interfering RNA (siRNA) Transfection
2.9. Quantitative Real-Time PCR (qRT–PCR)
2.10. Western Blot Analysis
2.11. Cell Proliferation Assay
2.12. Cell Migration and Invasion Assay
2.13. Statistical Analysis
3. Results
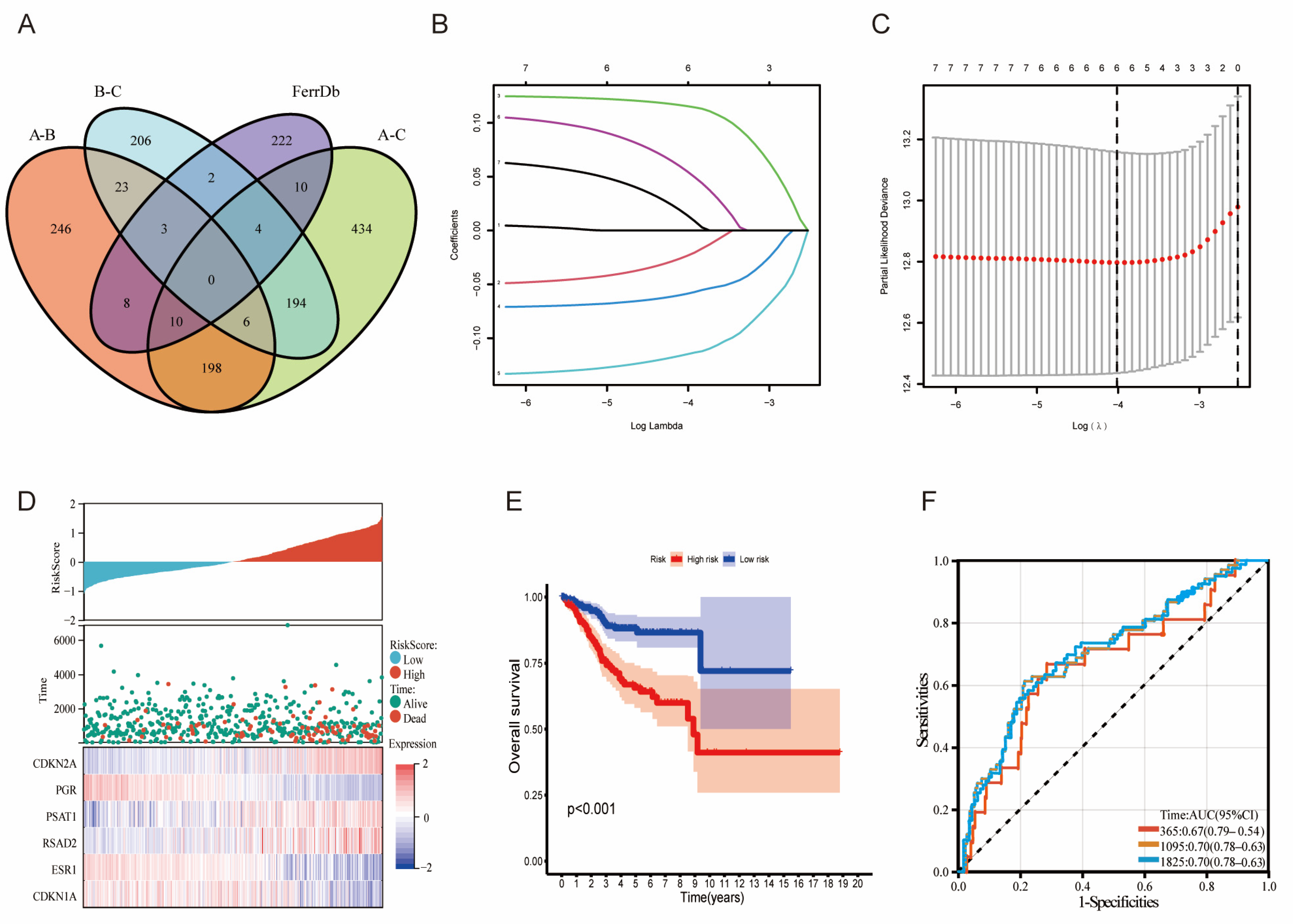
3.1. Identification and Clustering of LMRGs
3.2. Signature Construction Based on LMRGs and FAGs
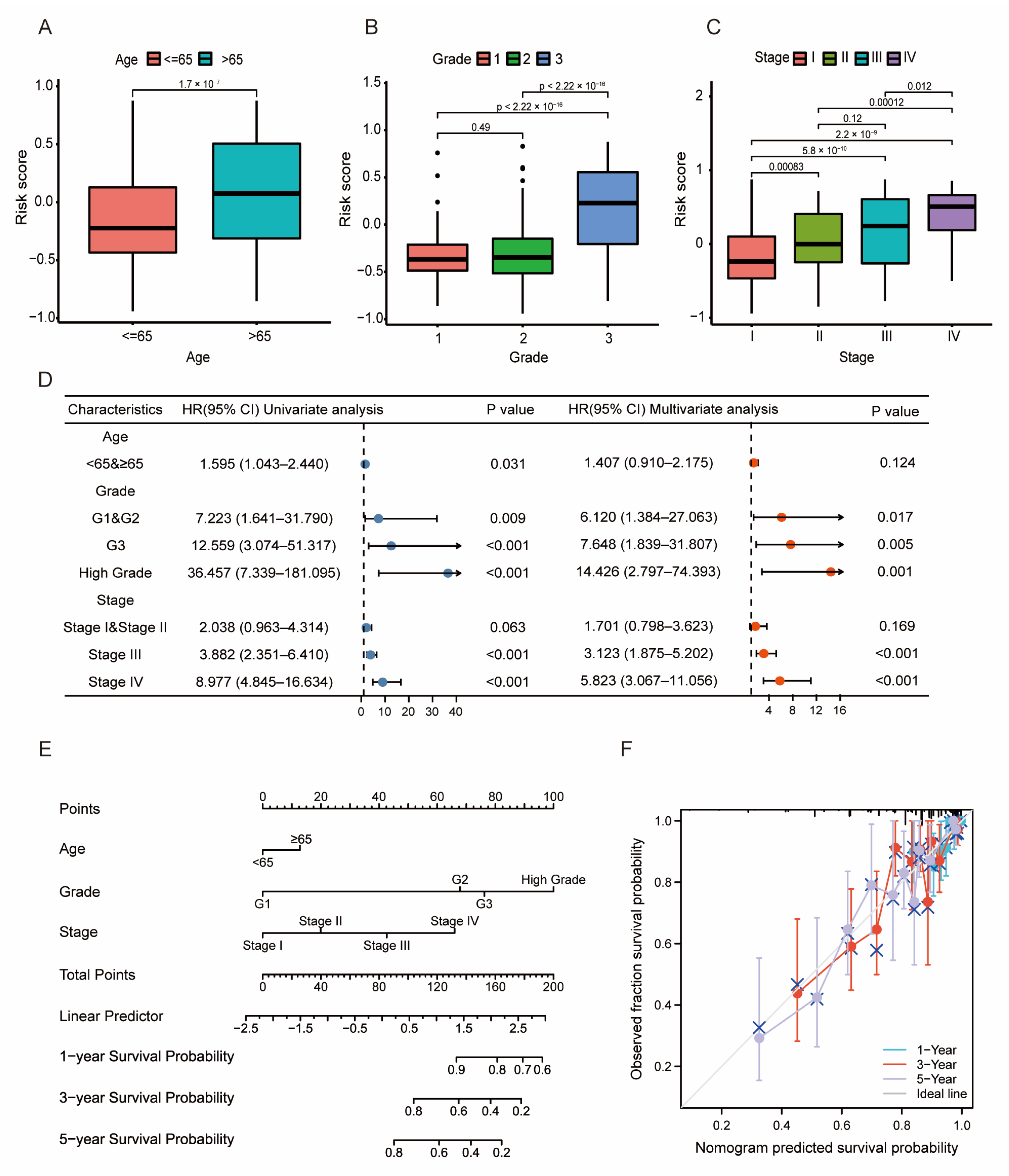
3.3. Prognosis and Validation of the LMRG- and FAG-Based Signature
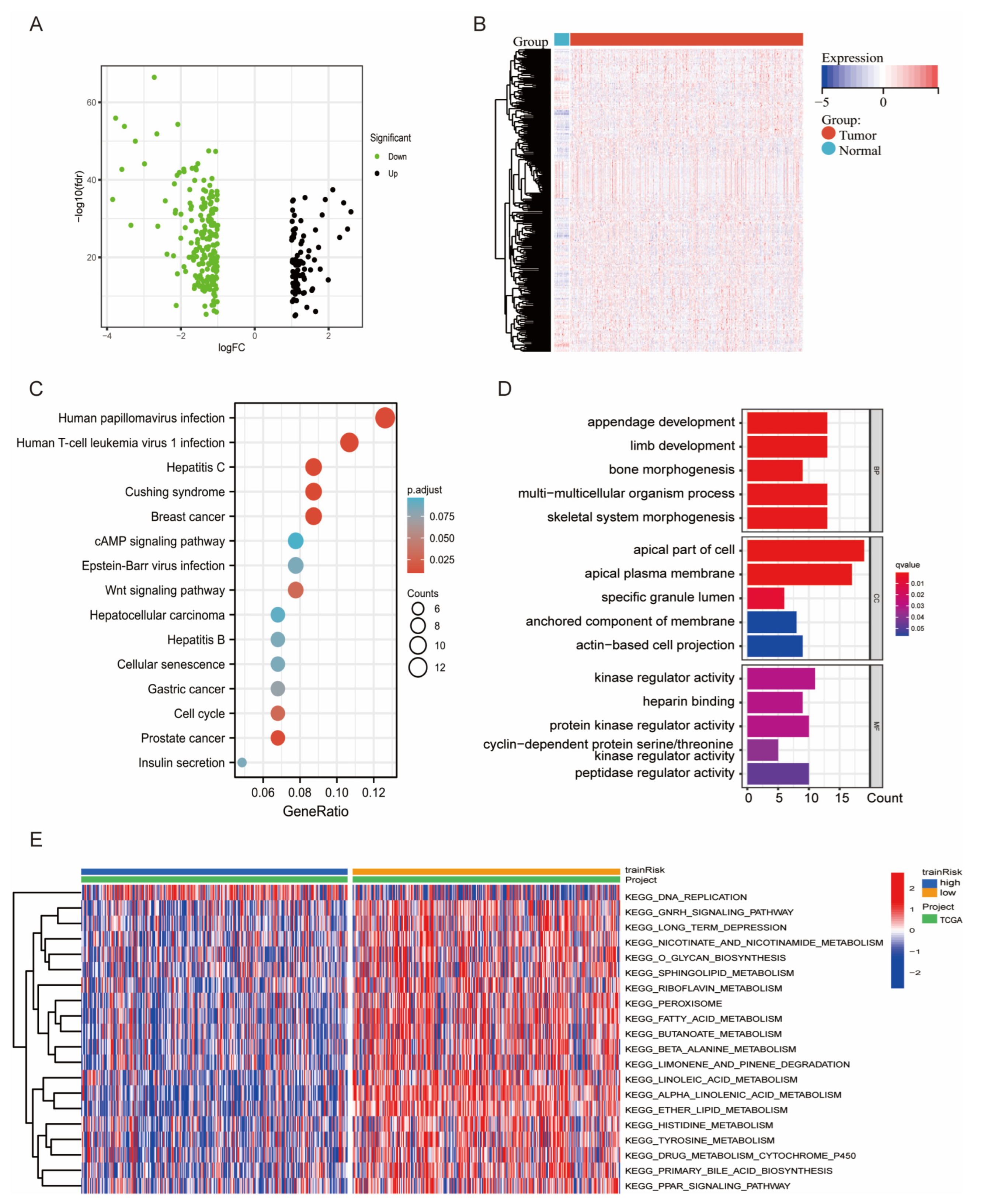
3.4. DEG and Functional Enrichment Analyses of the Signature
3.5. Relationship between the Tumor Mutational Burden (TMB) and the Risk Model
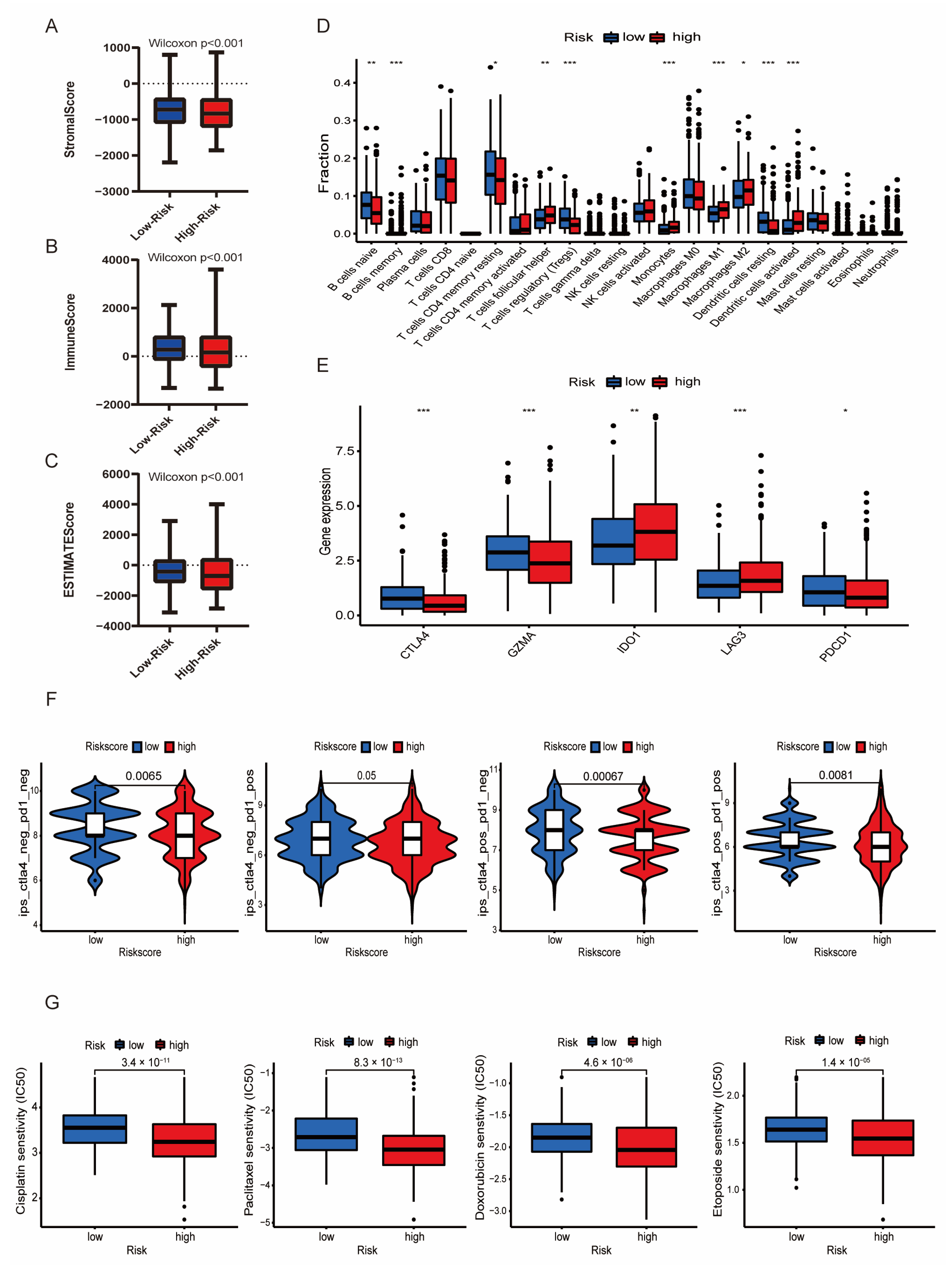
3.6. Immune Infiltration Associated with the LMRG-FAG-Based Signature
3.7. Immunotherapy and Chemotherapy in Different Risk Groups
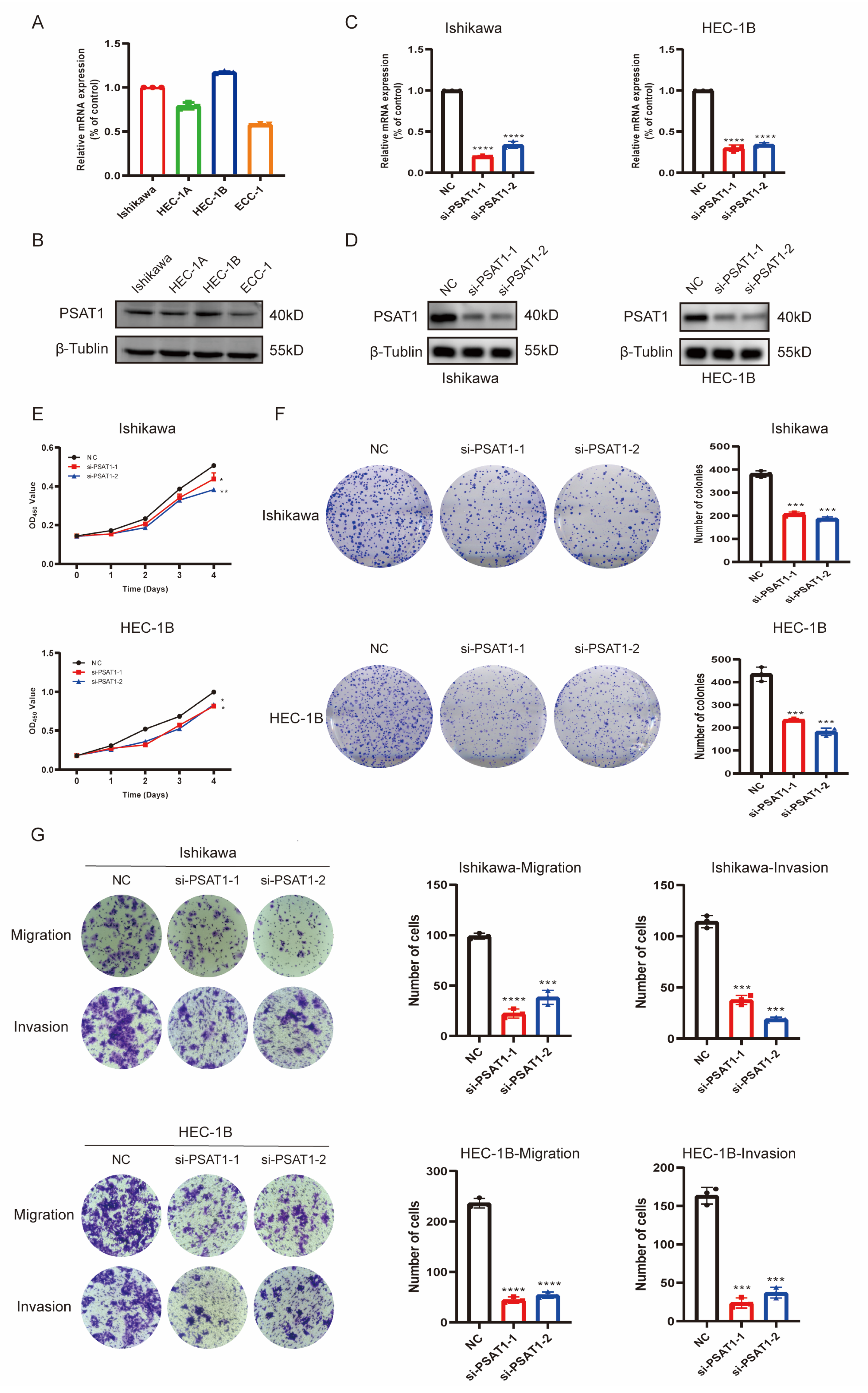
3.8. In Vitro Function of the Risk Gene PSAT1 in UCEC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Lu, K.H.; Broaddus, R.R. Endometrial Cancer. N. Engl. J. Med. 2020, 383, 2053–2064. [Google Scholar] [CrossRef]
- Miyakawa, K.; Jeremiah, S.S.; Ogawa, M.; Nishi, M.; Ohnishi, M.; Ryo, A. Crosstalk between the innate immune system and selective autophagy in hepatitis B virus infection. Autophagy 2022, 18, 2006–2007. [Google Scholar] [CrossRef] [PubMed]
- Noder, W. Usefulness of rest values of HMV in functional analytical studies. Z. Kreislaufforsch. 1969, 58, 1307–1314. [Google Scholar] [PubMed]
- Connor, E.V.; Rose, P.G. Management Strategies for Recurrent Endometrial Cancer. Expert Rev. Anticancer Ther. 2018, 18, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Voelker, R. Immunotherapy Approved for Endometrial Cancer. JAMA 2021, 325, 2143. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Ma, X.; Fischer, J.V.; Sun, C.; Kong, B.; Zhang, Q. Immunotherapy in endometrial cancer: Rationale, practice and perspectives. Biomark Res. 2021, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef] [PubMed]
- Onstad, M.A.; Schmandt, R.E.; Lu, K.H. Addressing the Role of Obesity in Endometrial Cancer Risk, Prevention, and Treatment. J. Clin. Oncol. 2016, 34, 4225–4230. [Google Scholar] [CrossRef]
- Fader, A.N.; Arriba, L.N.; Frasure, H.E.; von Gruenigen, V.E. Endometrial cancer and obesity: Epidemiology, biomarkers, prevention and survivorship. Gynecol. Oncol. 2009, 114, 121–127. [Google Scholar] [CrossRef]
- Tan, X.; Liu, S.; Yao, L.; Cui, G.; Liu, J.; Ding, J. Comprehensive Analysis of a Novel Lipid Metabolism-Related Gene Signature for Predicting the Prognosis and Immune Landscape in Uterine Corpus Endometrial Carcinoma. J. Oncol. 2022, 2022, 8028825. [Google Scholar] [CrossRef]
- Xu, H.; Ye, D.; Ren, M.; Zhang, H.; Bi, F. Ferroptosis in the tumor microenvironment: Perspectives for immunotherapy. Trends Mol. Med. 2021, 27, 856–867. [Google Scholar] [CrossRef]
- Lane, D.J.R.; Metselaar, B.; Greenough, M.; Bush, A.I.; Ayton, S.J. Ferroptosis and NRF2: An emerging battlefield in the neurodegeneration of Alzheimer’s disease. Essays Biochem. 2021, 65, 925–940. [Google Scholar] [CrossRef]
- Alim, I.; Caulfield, J.T.; Chen, Y.; Swarup, V.; Geschwind, D.H.; Ivanova, E.; Seravalli, J.; Ai, Y.; Sansing, L.H.; Ste Marie, E.J.; et al. Selenium Drives a Transcriptional Adaptive Program to Block Ferroptosis and Treat Stroke. Cell 2019, 177, 1262–1279.e1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wu, Y.; Chen, S.; Hou, M.; Yang, Y.; Xie, M. Construction and Validation of a Ferroptosis-Related Prognostic Model for Endometrial Cancer. Front. Genet. 2021, 12, 729046. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Meng, H.; Yin, Y.; Zhu, H.; Ni, T. Identification of the Prognostic Signature Associated with Tumor Immune Microenvironment of Uterine Corpus Endometrial Carcinoma Based on Ferroptosis-Related Genes. Front. Cell Dev. Biol. 2021, 9, 735013. [Google Scholar] [CrossRef] [PubMed]
- Weijiao, Y.; Fuchun, L.; Mengjie, C.; Xiaoqing, Q.; Hao, L.; Yuan, L.; Desheng, Y. Immune infiltration and a ferroptosis-associated gene signature for predicting the prognosis of patients with endometrial cancer. Aging 2021, 13, 16713–16732. [Google Scholar] [CrossRef]
- Gao, M.; Yi, J.; Zhu, J.; Minikes, A.M.; Monian, P.; Thompson, C.B.; Jiang, X. Role of Mitochondria in Ferroptosis. Mol. Cell 2019, 73, 354–363 e353. [Google Scholar] [CrossRef] [Green Version]
- Jelinek, A.; Heyder, L.; Daude, M.; Plessner, M.; Krippner, S.; Grosse, R.; Diederich, W.E.; Culmsee, C. Mitochondrial rescue prevents glutathione peroxidase-dependent ferroptosis. Free Radic. Biol. Med. 2018, 117, 45–57. [Google Scholar] [CrossRef]
- De Bortoli, M.; Taverna, E.; Maffioli, E.; Casalini, P.; Crisafi, F.; Kumar, V.; Caccia, C.; Polli, D.; Tedeschi, G.; Bongarzone, I. Lipid accumulation in human breast cancer cells injured by iron depletors. J. Exp. Clin. Cancer Res. 2018, 37, 75. [Google Scholar] [CrossRef] [Green Version]
- Cilloni, D.; Ravera, S.; Calabrese, C.; Gaidano, V.; Niscola, P.; Balleari, E.; Gallo, D.; Petiti, J.; Signorino, E.; Rosso, V.; et al. Iron overload alters the energy metabolism in patients with myelodysplastic syndromes: Results from the multicenter FISM BIOFER study. Sci. Rep. 2020, 10, 9156. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. A powerful cell-protection system prevents cell death by ferroptosis. Nature 2019, 575, 597–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doll, S.; Freitas, F.P.; Shah, R.; Aldrovandi, M.; da Silva, M.C.; Ingold, I.; Goya Grocin, A.; Xavier da Silva, T.N.; Panzilius, E.; Scheel, C.H.; et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019, 575, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Miess, H.; Dankworth, B.; Gouw, A.M.; Rosenfeldt, M.; Schmitz, W.; Jiang, M.; Saunders, B.; Howell, M.; Downward, J.; Felsher, D.W.; et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 2018, 37, 5435–5450. [Google Scholar] [CrossRef] [PubMed]
- Kapralov, A.A.; Yang, Q.; Dar, H.H.; Tyurina, Y.Y.; Anthonymuthu, T.S.; Kim, R.; St Croix, C.M.; Mikulska-Ruminska, K.; Liu, B.; Shrivastava, I.H.; et al. Redox lipid reprogramming commands susceptibility of macrophages and microglia to ferroptotic death. Nat. Chem. Biol. 2020, 16, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Kon, N.; Chen, D.; Li, T.; Liu, T.; Jiang, L.; Song, S.; Tavana, O.; Gu, W. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 2019, 21, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol. Biol. 2014, 1150, 153–160. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, H.; Deng, R.; Wang, Z.; Li, N.; Qi, Z.; Zhao, J.; Huang, W. Multi-omics analysis reveals prognostic value of tumor mutation burden in hepatocellular carcinoma. Cancer Cell Int. 2021, 21, 342. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Yu, W.; Lei, Q.; Yang, L.; Qin, G.; Liu, S.; Wang, D.; Ping, Y.; Zhang, Y. Contradictory roles of lipid metabolism in immune response within the tumor microenvironment. J. Hematol. Oncol. 2021, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Jiang, X. A Physiological Function for Ferroptosis in Tumor Suppression by the Immune System. Cell Metab. 2019, 30, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, S.; Jin, Z.; Shi, M. Lipid-mediated regulation of the cancer-immune crosstalk. Pharmacol. Res. 2020, 161, 105131. [Google Scholar] [CrossRef]
- Fan, F.; Liu, P.; Bao, R.; Chen, J.; Zhou, M.; Mo, Z.; Ma, Y.; Liu, H.; Zhou, Y.; Cai, X.; et al. A Dual PI3K/HDAC Inhibitor Induces Immunogenic Ferroptosis to Potentiate Cancer Immune Checkpoint Therapy. Cancer Res. 2021, 81, 6233–6245. [Google Scholar] [CrossRef] [PubMed]
- Thuesen, L.; Christiansen, J.S.; Mogensen, C.E.; Henningsen, P. Left ventricular wall mass and wall thickness in insulin dependent diabetic patients without clinical signs of ischaemic heart disease. Ann. Clin. Res. 1988, 20 (Suppl. 48), 7–9. [Google Scholar]
- Di Tucci, C.; Capone, C.; Galati, G.; Iacobelli, V.; Schiavi, M.C.; Di Donato, V.; Muzii, L.; Panici, P.B. Immunotherapy in endometrial cancer: New scenarios on the horizon. J. Gynecol. Oncol. 2019, 30, e46. [Google Scholar] [CrossRef] [Green Version]
- Amant, F.; Moerman, P.; Neven, P.; Timmerman, D.; Van Limbergen, E.; Vergote, I. Endometrial cancer. Lancet 2005, 366, 491–505. [Google Scholar] [CrossRef]
- Lee, Y.C.; Lheureux, S.; Oza, A.M. Treatment strategies for endometrial cancer: Current practice and perspective. Curr. Opin. Obstet. Gynecol. 2017, 29, 47–58. [Google Scholar] [CrossRef]
- Dessources, K.; Miller, K.M.; Kertowidjojo, E.; Da Cruz Paula, A.; Zou, Y.; Selenica, P.; da Silva, E.M.; Benayed, R.; Ashley, C.W.; Abu-Rustum, N.R.; et al. ESR1 hotspot mutations in endometrial stromal sarcoma with high-grade transformation and endocrine treatment. Mod. Pathol. 2022, 35, 972–978. [Google Scholar] [CrossRef]
- Rodriguez, A.C.; Vahrenkamp, J.M.; Berrett, K.C.; Clark, K.A.; Guillen, K.P.; Scherer, S.D.; Yang, C.H.; Welm, B.E.; Janat-Amsbury, M.M.; Graves, B.J.; et al. ETV4 Is Necessary for Estrogen Signaling and Growth in Endometrial Cancer Cells. Cancer Res. 2020, 80, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, Z.; Vahrenkamp, J.M.; Berrett, K.C.; Arnesen, S.; Gertz, J. Estrogen-independent molecular actions of mutant estrogen receptor 1 in endometrial cancer. Genome Res. 2019, 29, 1429–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Gao, W.; Zhang, Q.; Cheng, X.; Liu, Y.; Qi, Z.; Li, T. Ferroptosis and Autophagy-Related Genes in the Pathogenesis of Ischemic Cardiomyopathy. Front. Cardiovasc. Med. 2022, 9, 906753. [Google Scholar] [CrossRef]
- Cheng, T.; Wu, Y.; Liu, Z.; Yu, Y.; Sun, S.; Guo, M.; Sun, B.; Huang, C. CDKN2A-mediated molecular subtypes characterize the hallmarks of tumor microenvironment and guide precision medicine in triple-negative breast cancer. Front. Immunol. 2022, 13, 970950. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jia, H.; Huang, L.; Li, S.; Wang, C.; Aikemu, B.; Yang, G.; Hong, H.; Yang, X.; Zhang, S.; et al. An Original Ferroptosis-Related Gene Signature Effectively Predicts the Prognosis and Clinical Status for Colorectal Cancer Patients. Front. Oncol. 2021, 11, 711776. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.S.; Lee, J.H.; Jung, N.C.; Choi, S.Y.; Park, S.Y.; Yoo, J.Y.; Song, J.Y.; Seo, H.G.; Lee, H.S.; Lim, D.S. Rsad2 is necessary for mouse dendritic cell maturation via the IRF7-mediated signaling pathway. Cell Death Dis. 2018, 9, 823. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Song, C.; Wang, Y.; Lei, Z.; Xu, F.; Guan, H.; Chen, A.; Li, F. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal 2017, 34, 55–65. [Google Scholar] [CrossRef]
- Montrose, D.C.; Saha, S.; Foronda, M.; McNally, E.M.; Chen, J.; Zhou, X.K.; Ha, T.; Krumsiek, J.; Buyukozkan, M.; Verma, A.; et al. Exogenous and Endogenous Sources of Serine Contribute to Colon Cancer Metabolism, Growth, and Resistance to 5-Fluorouracil. Cancer Res. 2021, 81, 2275–2288. [Google Scholar] [CrossRef]
- Nwosu, Z.C.; Battello, N.; Rothley, M.; Pioronska, W.; Sitek, B.; Ebert, M.P.; Hofmann, U.; Sleeman, J.; Wolfl, S.; Meyer, C.; et al. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J. Exp. Clin. Cancer Res. 2018, 37, 211. [Google Scholar] [CrossRef]
- DeNicola, G.M.; Chen, P.H.; Mullarky, E.; Sudderth, J.A.; Hu, Z.; Wu, D.; Tang, H.; Xie, Y.; Asara, J.M.; Huffman, K.E.; et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015, 47, 1475–1481. [Google Scholar] [CrossRef] [Green Version]
- Chida, K.; Kawazoe, A.; Kawazu, M.; Suzuki, T.; Nakamura, Y.; Nakatsura, T.; Kuwata, T.; Ueno, T.; Kuboki, Y.; Kotani, D.; et al. A Low Tumor Mutational Burden and PTEN Mutations Are Predictors of a Negative Response to PD-1 Blockade in MSI-H/dMMR Gastrointestinal Tumors. Clin. Cancer Res. 2021, 27, 3714–3724. [Google Scholar] [CrossRef]
- Owada-Ozaki, Y.; Muto, S.; Takagi, H.; Inoue, T.; Watanabe, Y.; Fukuhara, M.; Yamaura, T.; Okabe, N.; Matsumura, Y.; Hasegawa, T.; et al. Prognostic Impact of Tumor Mutation Burden in Patients with Completely Resected Non-Small Cell Lung Cancer: Brief Report. J. Thorac. Oncol. 2018, 13, 1217–1221. [Google Scholar] [CrossRef] [Green Version]
- Byun, D.J.; Wolchok, J.D.; Rosenberg, L.M.; Girotra, M. Cancer immunotherapy—Immune checkpoint blockade and associated endocrinopathies. Nat. Rev. Endocrinol. 2017, 13, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Greenwald, R.J.; Boussiotis, V.A.; Lorsbach, R.B.; Abbas, A.K.; Sharpe, A.H. CTLA-4 regulates induction of anergy in vivo. Immunity 2001, 14, 145–155. [Google Scholar] [CrossRef]
- Topalian, S.L.; Taube, J.M.; Anders, R.A.; Pardoll, D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 2016, 16, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twomey, J.D.; Zhang, B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J. 2021, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, P.J.; Billingsley, C.C.; Lankes, H.A.; Ali, S.; Cohn, D.E.; Broaddus, R.J.; Ramirez, N.; Pritchard, C.C.; Hampel, H.; Chassen, A.S.; et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers from GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J. Clin. Oncol. 2015, 33, 4301–4308. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, P.; Lu, J.; Zhang, P.; Zhang, S. Comprehensive Analysis of Prognosis and Immune Landscapes Based on Lipid-Metabolism- and Ferroptosis-Associated Signature in Uterine Corpus Endometrial Carcinoma. Diagnostics 2023, 13, 870. https://doi.org/10.3390/diagnostics13050870
Yang P, Lu J, Zhang P, Zhang S. Comprehensive Analysis of Prognosis and Immune Landscapes Based on Lipid-Metabolism- and Ferroptosis-Associated Signature in Uterine Corpus Endometrial Carcinoma. Diagnostics. 2023; 13(5):870. https://doi.org/10.3390/diagnostics13050870
Chicago/Turabian StyleYang, Pusheng, Jiawei Lu, Panpan Zhang, and Shu Zhang. 2023. "Comprehensive Analysis of Prognosis and Immune Landscapes Based on Lipid-Metabolism- and Ferroptosis-Associated Signature in Uterine Corpus Endometrial Carcinoma" Diagnostics 13, no. 5: 870. https://doi.org/10.3390/diagnostics13050870





