Evaluation of the Microbiome Identification of Forensically Relevant Biological Fluids: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Volunteers and Setting
2.2. Sample Collection
2.3. Sample Preparation
2.4. Sample Extraction
2.5. Metagenomic Investigation
2.6. Statistical Analysis
3. Results
3.1. Characteristic of Volunteers
3.2. Oral Microbiome Analysis
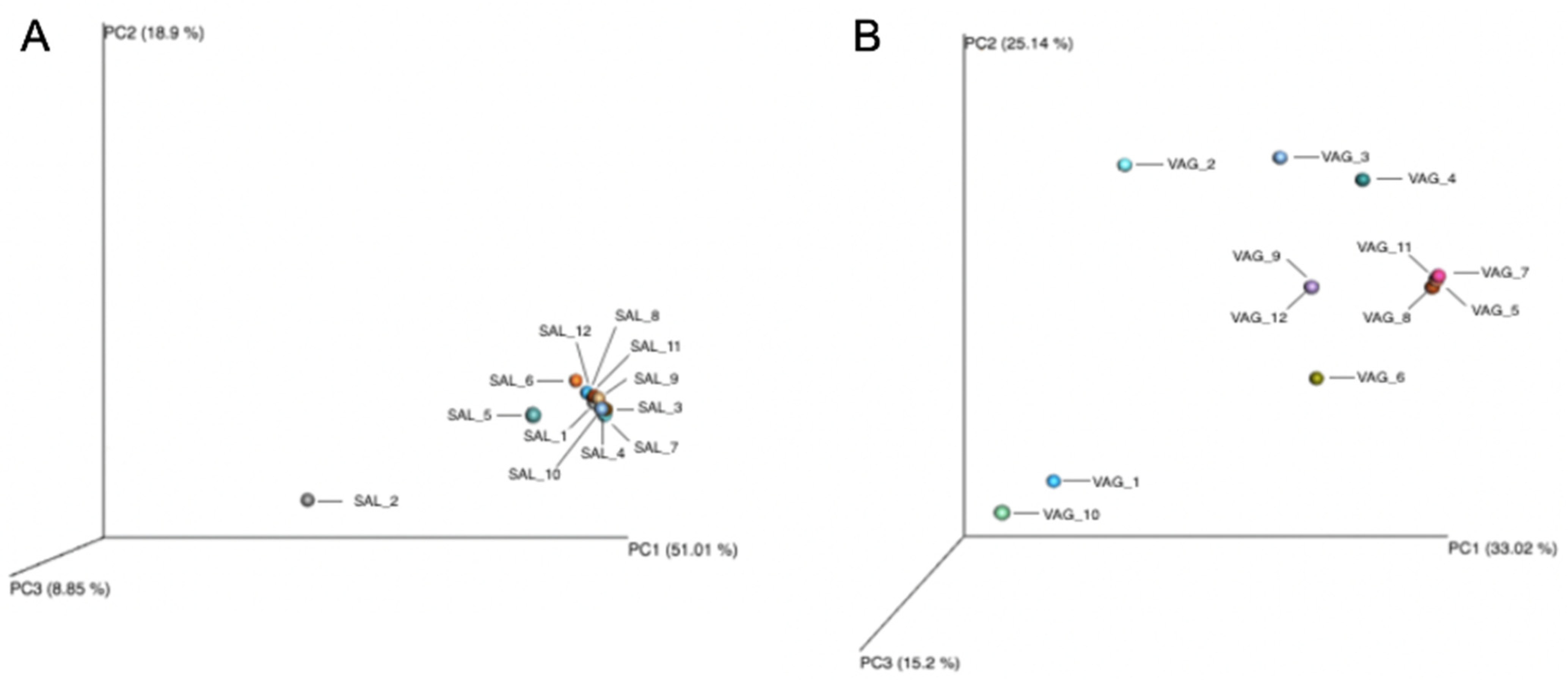
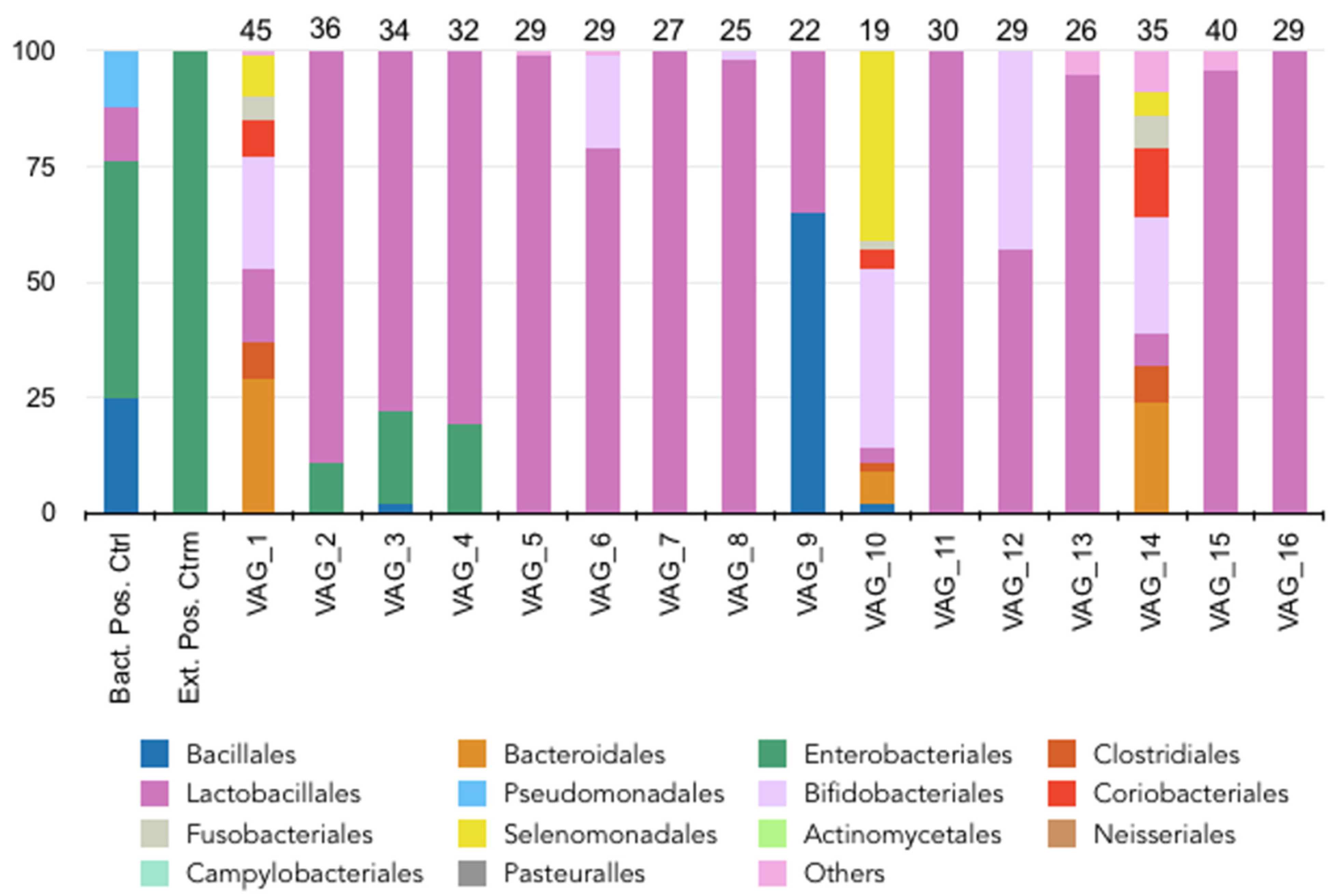
3.3. Vaginal Microbiome
3.4. Other Biological Fluids
3.5. Biological Fluid Mixture Assessment
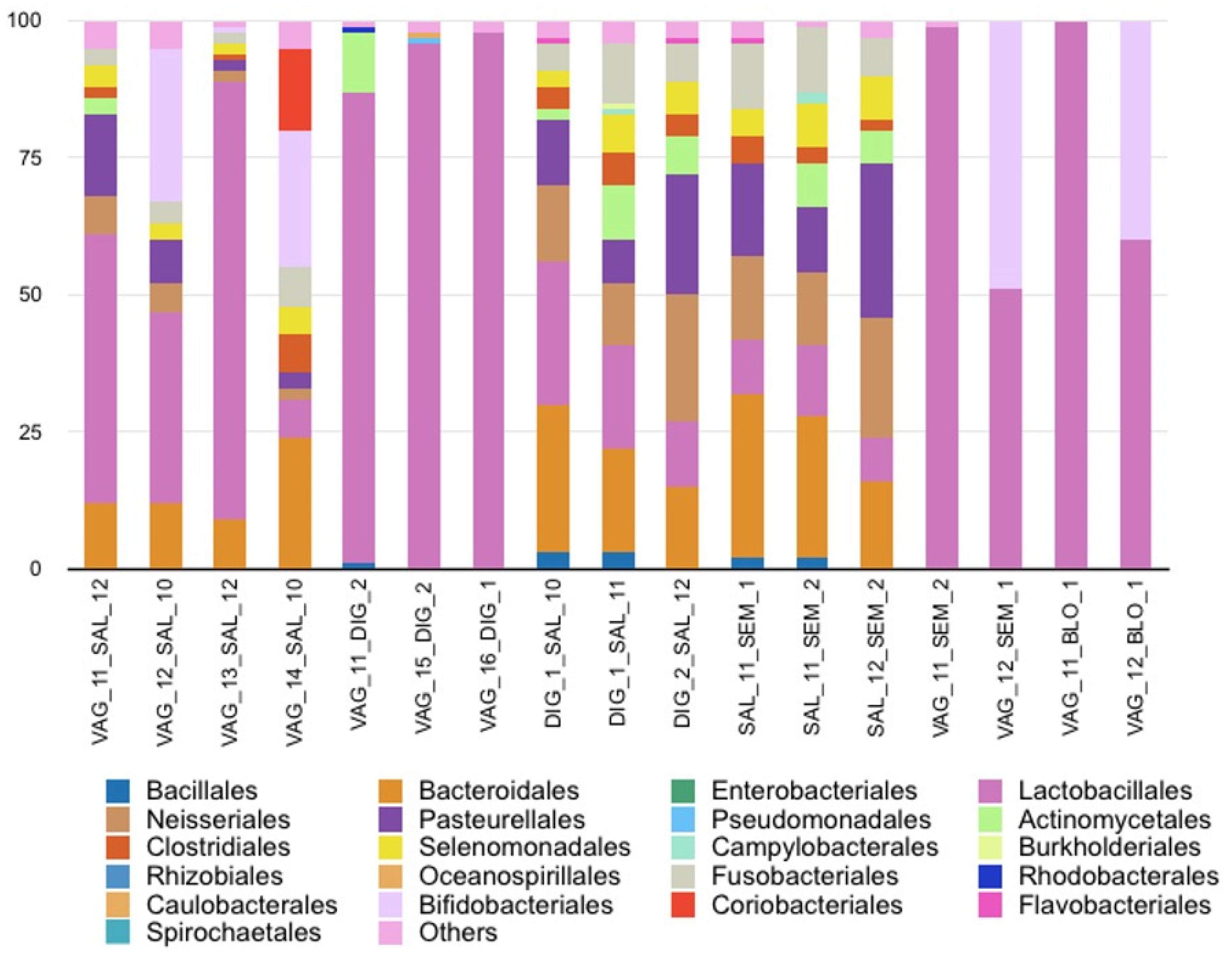
3.5.1. Saliva and Vaginal Fluid Mixtures
3.5.2. Vaginal Fluid and Digital Sample Mixtures
3.5.3. Saliva and Digital Sample Mixtures
3.5.4. Saliva and Semen Mixtures
3.5.5. Vaginal Fluid and Semen Mixtures
3.5.6. Vaginal Fluid and Blood Mixtures
3.5.7. Beta Diversity among the Fluid Mixtures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popkin, B.M.; D’Anci, K.E.; Rosenberg, I.H. Water, hydration, and health. Nutr. Rev. 2010, 68, 439–458. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Gouello, A.; Dunyach-Remy, C.; Siatka, C.; Lavigne, J.P. Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics 2021, 12, 1. [Google Scholar] [CrossRef]
- Sijen, T.; Harbison, S. On the Identification of Body Fluids and Tissues: A Crucial Link in the Investigation and Solution of Crime. Genes 2021, 12, 1728. [Google Scholar] [CrossRef]
- Harbison, S.; Fleming, R. Forensic body fluid identification: State of the art. Res. Rep. Forensic Med. Sc. 2016, 6, 11–23. [Google Scholar] [CrossRef]
- Dobay, A.; Haas, C.; Fucile, G.; Downey, N.; Morrison, H.G.; Kratzer, A.; Arora, N. Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci. Int. Genet. 2019, 40, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Han, X.; Guan, T.; Wang, Z.; Zhang, S.; Liu, C.; Liu, C.; Chen, L. Effect of indoor environmental exposure on seminal microbiota and its application in body fluid identification. Forensic Sci. Int. 2020, 314, 110417. [Google Scholar] [CrossRef]
- Yao, T.; Han, X.; Guan, T.; Zhai, C.; Liu, C.; Liu, C.; Zhu, B.; Chen, L. Exploration of the microbiome community for saliva, skin, and a mixture of both from a population living in Guangdong. Int. J. Leg. Med. 2021, 135, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Sguazzi, G.; Mickleburgh, H.L.; Ghignone, S.; Voyron, S.; Renò, F.; Migliario, M.; Sellitto, F.; Lovisolo, F.; Camurani, G.; Ogbanga, N.; et al. Microbial DNA in human nucleic acid extracts: Recoverability of the microbiome in DNA extracts stored frozen long-term and its potential and ethical implications for forensic investigation. Forensic Sci. Int. Genet. 2022, 59, 102686. [Google Scholar] [CrossRef]
- Adserias-Garriga, J.; Quijada, N.M.; Hernandez, M.; Rodríguez Lazaro, D.; Steadman, D.; Garcia-Gil, L.J. Dynamics of the oral microbiota as a tool to estimate time since death. Mol. Oral. Microbiol. 2017, 32, 511–516. [Google Scholar] [CrossRef]
- Belk, A.; Xu, Z.Z.; Carter, D.O.; Lynne, A.; Bucheli, S.; Knight, R.; Metcalf, J.L. Microbiome Data accurately predicts the postmortem interval using random forest regression models. Genes 2018, 9, 10. [Google Scholar] [CrossRef]
- Ghemrawi, M.; Torres, A.R.; Duncan, G.; Colwell, R.; Dadlani, M.; McCord, B. The genital microbiome and its potential for detecting sexual assault. Forensic Sci. Int. Genet. 2021, 51, 102432. [Google Scholar] [CrossRef] [PubMed]
- Ogbanga, N.; Nelson, A.; Ghignone, S.; Voyron, S.; Lovisolo, F.; Sguazzi, G.; Renò, F.; Migliario, M.; Gino, S.; Procopio, N. The Oral Microbiome for Geographic Origin: An Italian Study. Forensic Sci. Int. Genet. 2023, 64, 102841. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Clarke, T.; Brinkac, L.; Greco, C.; Nelson, K.E. Forensic microbiome database: A tool for forensic geolocation meta-analysis using publicly available 16S rRNA microbiome sequencing. Front. Microbiol. 2012, 12, 644861. [Google Scholar] [CrossRef]
- Anyaso-Samuel, S.; Sachdeva, A.; Guha, S.; Datta, S. Metagenomic geolocation prediction using an adaptive ensemble classifier. Front. Genet. 2021, 12, 642282. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.H.; Gomez, A.; Singh, H.; Nelson, K.E.; Brinkac, L.M. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 2017, 30, 141–147. [Google Scholar] [CrossRef]
- Budowle, B.; Connell, N.D.; Bielecka-Oder, A.; Colwell, R.R.; Corbett, C.R.; Fletcher, J.; Forsman, M.; Kadavy, D.R.; Markotic, A.; Morse, S.A.; et al. Validation of high throughput sequencing and microbial forensics applications. Investig. Genet. 2014, 5, 9. [Google Scholar] [CrossRef]
- Díez López, C.; Montiel González, D.; Haas, C.; Vidaki, A.; Kayser, M. Microbiome-based body site of origin classification of forensically relevant blood traces. Forensic Sci. Int. Genet. 2020, 47, 102280. [Google Scholar] [CrossRef]
- Volpellier, M.; Hirve, R.; Duckett, C. Genital injuries and allegation of digital vaginal penetration—A retrospective examination of forensic case notes. J. Forensic Leg. Med. 2021, 80, 102154. [Google Scholar] [CrossRef]
- Mullen, C.M.; Aneja, K.K.; Beyhan, S.; Cho, C.E.; Woloszynek, S.; Convertino, M.; McCoy, S.J.; Zhang, Y.; Anderson, M.Z.; Alvarez-Ponce, D.; et al. Emerging Priorities for Microbiome Research. Front. Microbiol. 2020, 11, 136. [Google Scholar]
- Plé, C.; Vandenborght, L.E.; Adele-Dit-Renseville, N.; Breton, J.; Daniel, C.; Ferreira, S.; Benoît, F. Snapshot on a Pilot Metagenomic Study for the Appraisal of Gut Microbial Diversity in Mice, Cat, and Man. Gastroenterol. Res. Pract. 2016, 2016, 6587825. [Google Scholar]
- Zaura, E.; Keijser, B.J.F.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.O.; Coutinho-Filho, W.P.; Pimenta, F.P.; Pereira, G.A.; Pereira, J.A.A.; Mattos-Guaraldi, A.L.; Hirata, R., Jr. Periodontal disease as reservoir for multi-resistant and hydrolytic enterobacterial species. Lett. Appl. Microbiol. 2007, 44, 488–494. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Stewart, E.M.; Mylotte, J.M. Colonization of dental plaque by respiratory pathogens in medical intensive care patients. Crit. Care Med. 1992, 20, 740–745. [Google Scholar] [CrossRef]
- Chopyk, J.; Bojanowski, C.M.; Shin, J.; Moshensky, A.; Fuentes, A.L.; Bonde, S.S.; Chuki, D.; Pride, D.T.; Alexander, L.E. Compositional Differences in the Oral Microbiome of E-cigarette Users. Front. Microbiol. 2021, 12, 599664. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peters, B.A.; Dominianni, C.; Zhang, Y.; Pei, Z.; Yang, L.; Ma, Y.; Purdue, M.P.; Jacobs, E.J.; Gapstur, S.M.; et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016, 10, 2435–2446. [Google Scholar] [CrossRef]
- Kennel de March, A.; Béné, M.C.; Derniame, S.; Massin, F.; Aguilar, P.; Faure, G. Tabac et immunité muqueuse: Inflammation ou déficit immunitaire acquis. Rev. Francophone Lab. 2004, 361, 27–31. [Google Scholar]
- Chattopadhyay, S.; Ramachandran, P.; Malayil, L.; Mongodin, E.F.; Sapkota, A.R. Conventional tobacco products harbor unique and heterogenous microbiomes. Environ. Res. 2023, 220, 115205. [Google Scholar] [CrossRef]
- Park, B.; Koh, H.; Patatanian, M.; Reyes-Caballero, H.; Zhao, N.; Meinert, J.; Holbrook, J.T.; Leinbach, L.I.; Biswal, S. The mediating roles of the oral microbiome in saliva and subgingival sites between e-cigarette smoking and gingival inflammation. BMC Microbiol. 2023, 23, 35. [Google Scholar] [CrossRef]
- Kumar, P.S.; Matthews, C.R.; Joshi, V.; de Jager, M.; Aspiras, M. Tobacco Smoking Affects Bacterial Acquisition and Colonization in Oral Biofilms. Infect. Immun. 2011, 79, 4730–4738. [Google Scholar] [CrossRef]
- Hall, M.W.; Singh, N.; Ng, N.F.; Lam, D.K.; Goldberg, M.B.; Tenenbaum, H.C.; Neufeld, J.D.; Beiko, R.G.; Senadheera, D.B. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes 2017, 3, 2. [Google Scholar] [CrossRef]
- Kwon, M.S.; Lee, H.K. Host and Microbiome Interplay Shapes the Vaginal Microenvironment. Front. Immunol. 2022, 13, 919728. [Google Scholar] [CrossRef]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli Dominance and Vaginal pH: Why Is the Human Vaginal Microbiome Unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef]
- Aldunate, M.; Srbinovski, D.; Hearps, A.C.; Latham, C.F.; Ramsland, P.A.; Gugasyan, R.; Cone, R.A.; Tachedjian, G. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front. Physiol. 2015, 6, 164. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Y.; Chen, T.; Li, R. The Female Vaginal Microbiome in Health and Bacterial Vaginosis. Front. Cell Infect. Microbiol. 2021, 11, 631972. [Google Scholar] [CrossRef]
- Kenetta, L.N.; Forney, L.J. Unraveling the Dynamics of the Human Vaginal Microbiome, Yale. J. Biol. Med. 2016, 89, 331–337. [Google Scholar]
- Romero, R.; Hassan, S.S.; Gajer, P.; Tarca, A.L.; Fadrosh, D.W.; Nikita, L.; Galuppi, M.; Lamont, R.F.; Chaemsaithong, P.; Miranda, J.; et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014, 2, 4. [Google Scholar]
- Kim, S.; Seo, H.; Rahim, M.D.A.; Lee, S.; Kim, Y.S.; Song, H.Y. Changes in the Microbiome of Vaginal Fluid after Menopause in Korean Women. J. Microbiol. Biotechnol. 2021, 31, 1490–1500. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Dass, M.; Singh, Y.; Ghai, M.A. A Review on Microbial Species for Forensic Body Fluid Identification in Healthy and Diseased Humans. Curr. Microbiol. 2023, 80, 299. [Google Scholar] [CrossRef] [PubMed]
- Procopio, N.; Lovisolo, F.; Sguazzi, G.; Ghignone, S.; Voyron, S.; Migliario, M.; Reno, F.; Sellitto, F.; D’Angiolella, G.; Tozzo, P.; et al. “Touch microbiome” as a potential tool for forensic investigation: A pilot study. J. Forensic Leg. Med. 2021, 82, 102223. [Google Scholar] [CrossRef] [PubMed]
- Ceccarani, C.; Foschi, C.; Parolin, C.; D’Antuono, A.; Gaspari, V.; Consolandi, C.; Laghi, L.; Camboni, T.; Vitali, B.; Severgnini, M.; et al. Diversity of vaginal microbiome and metabolome during genital infections. Sci. Rep. 2019, 9, 14095. [Google Scholar] [CrossRef] [PubMed]



| Samples | Number of Volunteers | ||||
|---|---|---|---|---|---|
| Saliva n = 12 | Vaginal n = 16 | Digital n = 2 | Semen n = 2 | Blood n = 1 | |
| Age, median (range) | 35 (26–50) | 29 (19–45) | 37 (24–50) | 37 (27–46) | 50 |
| Sex, women/men | 6/6 | 16/0 | 1/1 | 0/2 | 0/1 |
| Pregnancy | 1 | 0 | 0 | 0 | 0 |
| Smoking habits | 4 | 0 | 0 | 0 | 0 |
| E-cigarette habits | 1 | 0 | 0 | 0 | 0 |
| Microbiomes | |||||
|---|---|---|---|---|---|
| Oral | Vaginal | Semen | Digital Skin | Blood | |
| OTU 1 species, average | 46,752 | 8182 | 47,311 | 67,209 | 3800 |
| OTU species, standard deviation | 14,209 | 1991 | 6732 | 21,536 | 0791 |
| Shannon index, average | 2.99 | 1.02 | 2.78 | 2.98 | 1.64 |
| Shannon index, standard deviation | 0.33 | 0.06 | 0.26 | 0.68 | 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gouello, A.; Henry, L.; Chadli, D.; Salipante, F.; Gibert, J.; Boutet-Dubois, A.; Lavigne, J.-P. Evaluation of the Microbiome Identification of Forensically Relevant Biological Fluids: A Pilot Study. Diagnostics 2024, 14, 187. https://doi.org/10.3390/diagnostics14020187
Gouello A, Henry L, Chadli D, Salipante F, Gibert J, Boutet-Dubois A, Lavigne J-P. Evaluation of the Microbiome Identification of Forensically Relevant Biological Fluids: A Pilot Study. Diagnostics. 2024; 14(2):187. https://doi.org/10.3390/diagnostics14020187
Chicago/Turabian StyleGouello, Audrey, Laura Henry, Djamel Chadli, Florian Salipante, Joséphine Gibert, Adeline Boutet-Dubois, and Jean-Philippe Lavigne. 2024. "Evaluation of the Microbiome Identification of Forensically Relevant Biological Fluids: A Pilot Study" Diagnostics 14, no. 2: 187. https://doi.org/10.3390/diagnostics14020187





