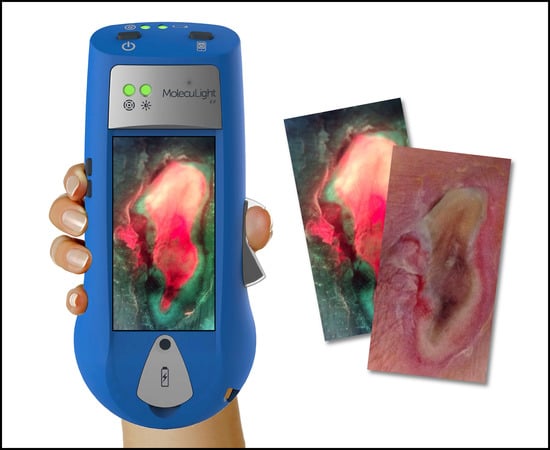
Understanding Real-Time Fluorescence Signals from Bacteria and Wound Tissues Observed with the MolecuLight i:XTM
Abstract
:1. Introduction
2. How Does the MolecuLight i:X Work?
3. Evidence for Bacterial Detection
4. Image Interpretation
4.1. Tissue Fluorescence
4.2. Bacterial Fluorescence
5. Methods to Minimize Imaging Artifacts and Misinterpretation
6. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, A. The wound infection continuum and its application to clinical practice. Ostomy Wound Manag. 2003, 49, 1–7. [Google Scholar]
- Rahim, K.; Saleha, S.; Zhu, X.; Huo, L.; Basit, A.; Franco, O.L. Bacterial Contribution in Chronicity of Wounds. Microb. Ecol. 2017, 73, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.; Sanford, N.; Gabrilska, R.; Oates, J.L.; Wilkinson, J.E.; Rumbaugh, K.P. Microbiota is a primary cause of pathogenesis of chronic wounds. J. Wound Care 2016, 25, S33–S43. [Google Scholar] [CrossRef] [PubMed]
- Bowler, P.G. The 10(5) bacterial growth guideline: Reassessing its clinical relevance in wound healing. Ostomy Wound Manag. 2003, 49, 44–53. [Google Scholar]
- Wilcox, J.R.; Carter, M.J.; Covington, S. Frequency of debridements and time to heal: A retrospective cohort study of 312 744 wounds. JAMA Dermatol. 2013, 149, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Schaper, N.C. Diabetic foot ulcer classification system for research purposes: A progress report on criteria for including patients in research studies. Diabetes Metab. Res. Rev. 2004, 20, S90–S95. [Google Scholar] [CrossRef] [PubMed]
- Kallstrom, G. Are quantitative bacterial wound cultures useful? J. Clin. Microbiol. 2014, 52, 2753–2756. [Google Scholar] [CrossRef] [PubMed]
- Copeland-Halperin, L.R.; Kaminsky, A.J.; Bluefeld, N.; Miraliakbari, R. Sample procurement for cultures of infected wounds: A systematic review. J. Wound Care 2016, 25, S4–S10. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, A.; Winfield-Davies, S. Audit of wound swab sampling: Why protocols could improve practice. Prof. Nurse 2003, 18, 338–343. [Google Scholar] [PubMed]
- Gardner, S.E.; Hillis, S.L.; Frantz, R.A. Clinical signs of infection in diabetic foot ulcers with high microbial load. Biol. Res. Nurs. 2009, 11, 119–128. [Google Scholar] [CrossRef] [PubMed]
- International Wound Infection Institute. IWII, Wound Infection in Clinical Practice; Wounds International: London, UK, 2016. [Google Scholar]
- Reddy, M.; Gill, S.S.; Wu, W.; Kalkar, S.R.; Rochon, P.A. Does this patient have an infection of a chronic wound? JAMA 2012, 307, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Sibbald, R.G.; Woo, K.; Ayello, E.A. Increased bacterial burden and infection: The story of NERDS and STONES. Adv. Skin Wound Care 2006, 19, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Serena, T.E.; Hanft, J.R.; Snyder, R. The lack of reliability of clinical examination in the diagnosis of wound infection: Preliminary communication. Int. J. Low Extrem. Wounds 2008, 7, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, E.; Jeffery, S.L.A. The Use of the MolecuLight i:X in Managing Burns: A Pilot Study. J. Burn Care Res. 2018, 39, 154–161. [Google Scholar] [PubMed]
- DaCosta, R.S.; Kulbatski, I.; Lindvere-Teene, L.; Starr, D.; Blackmore, K.; Silver, J.I.; Opoku, J.; Wu, Y.C.; Medeiros, P.J.; et al. Point-of-care autofluorescence imaging for real-time sampling and treatment guidance of bioburden in chronic wounds: First-in-human results. PLoS ONE 2015, 10, e0116623. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Rennie, M.Y.; Douglas, J. Using Bacterial Fluorescence Imaging and Antimicrobial Stewardship to Guide Wound Management Practices: A Case Series. Ostomy Wound Manag. 2018, 64, 18–28. [Google Scholar] [CrossRef]
- Ottolino-Perry, K.; Chamma, E.; Blackmore, K.M.; Lindvere-Teene, L.; Starr, D.; Tapang, K.; Rosen, C.F.; Pitcher, B.; Panzarella, T.; Linden, R.; et al. Improved detection of clinically relevant wound bacteria using autofluorescence image-guided sampling in diabetic foot ulcers. Int. Wound J. 2017, 14, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.Y.; Lindvere-Teene, L.; Tapang, K.; Linden, R. Point-of-care fluorescence imaging predicts the presence of pathogenic bacteria in wounds: A clinical study. J. Wound Care 2017, 26, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Kulbatski, I.; Medeiros, P.J.; Maeda, A.; Bu, J.; Xu, L.; Chen, Y.; DaCosta, R.S. Autofluorescence imaging device for real-time detection and tracking of pathogenic bacteria in a mouse skin wound model: Preclinical feasibility studies. J. Biomed. Opt. 2014, 19, 085002. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Smith, M.; Chu, A.; Lindvere-Teene, L.; Starr, D.; Tapang, K.; Shekhman, R.; Wong, O.; Linden, R.; DaCosta, R.S. Handheld fluorescence imaging device detects subclinical wound infection in an asymptomatic patient with chronic diabetic foot ulcer: A case report. Int. Wound J. 2016, 13, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, E.; Jeffery, S. Autofluorescence Imaging for Evaluating Debridement in Military and Trauma Wounds. Mil Med. 2018, 183, 429–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monici, M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 2005, 11, 227–256. [Google Scholar] [PubMed]
- Andersson-Engels, S.; af Klinteberg, C.; Svanberg, K.; Svanberg, S. In vivo fluorescence imaging for tissue diagnostics. Phys. Med. Biol. 1997, 42, 815–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGinley, K.J.; Webster, G.F.; Leyden, J.J. Facial follicular porphyrin fluorescence: Correlation with age and density of Propionibacterium acnes. Br. J. Dermatol. 1980, 102, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Rimington, C. Spectral-absorption coefficients of some porphyrins in the Soret-band region. Biochem. J. 1960, 75, 4. [Google Scholar] [CrossRef]
- Sliney, D.H. Optical radiation safety of medical light sources. Phys. Med. Biol. 1997, 42, 981–996. [Google Scholar] [CrossRef] [PubMed]
- Uttamlal, M.; Holmes-Smith, A.S. The excitation wavelength dependent fluorescence of porphyrins. Chem. Phys. Lett. 2008, 454, 6. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Datta, R.; Gratton, E.; Hochbaum, A.I. Metabolic fingerprinting of bacteria by fluorescence lifetime imaging microscopy. Sci. Rep. 2017, 7, 3743. [Google Scholar] [CrossRef] [PubMed]
- Dartnell, L.R.; Roberts, T.A.; Moore, G.; Ward, J.M.; Muller, J.P. Fluorescence characterization of clinically-important bacteria. PLoS ONE 2013, 8, e75270. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, N.L.; Tzeranis, D.S.; Wang, Y.; So, P.T.; Newman, D. Quantifying the dynamics of bacterial secondary metabolites by spectral multiphoton microscopy. ACS Chem. Biol. 2011, 6, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Kjeldstad, B.; Johnsson, A.; Sandberg, S. Influence of pH on porphyrin production in Propionibacterium acnes. Arch. Dermatol. Res. 1984, 276, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.L.; Shalita, A.R.; Poh-Fitzpatrick, M.B. Comparative studies of porphyrin production in Propionibacterium acnes and Propionibacterium granulosum. J. Bacteriol. 1978, 133, 811–815. [Google Scholar] [PubMed]
- Philipp-Dormston, W.K.; Doss, M. Comparison of porphyrin and heme biosynthesis in various heterotrophic bacteria. Enzyme 1973, 16, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.A.; Abdallah, M.A. The Fluorescent Pigment of Pseudomonas fluorescens: Biosynthesis, Purification and Physicochemical Properties. Microbiology 1978, 107, 9. [Google Scholar] [CrossRef]
- Dietel, W.; Pottier, R.; Pfister, W.; Schleier, P.; Zinner, K. 5-Aminolaevulinic acid (ALA) induced formation of different fluorescent porphyrins: A study of the biosynthesis of porphyrins by bacteria of the human digestive tract. J. Photochem. Photobiol. B 2007, 86, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Guillon, L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: Implications for metal homeostasis. Environ. Microbiol. 2013, 15, 1661–1673. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Huband, M.D.; Castanheira, M.; Flamm, R.K. Pseudomonas aeruginosa Antimicrobial Susceptibility Results from Four Years (2012 to 2015) of the International Network for Optimal Resistance Monitoring Program in the United States. Antimicrob. Agents Chemother. 2017, 61, e02252-16. [Google Scholar] [CrossRef] [PubMed]
- Blackshaw, E.L.; Jeffery, S.L.A. Efficacy of an imaging device at identifying the presence of bacteria in wounds at a plastic surgery outpatients clinic. J. Wound Care 2018, 27, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Hurley, C.M.; McClusky, P.; Sugrue, R.; Clover, A.J.; Kelly, E.J. The Efficacy of the MolecuLight i:X Wound Intelligence Device in the Outpatient Wound Care Clinic: A Pilot Study. J. Wound Care 2019. (In press) [Google Scholar]
- Raizman, R. Prospective clinical evaluation of fluorescence imaging in positively predicting the presence of Pseudomonas aeruginosa in chronic wounds. In Proceedings of the Annual European Wound Management Association Conference, Krakow, Poland, 9–11 May 2018. [Google Scholar]
- Unal, S.; Ersoz, G.; Demirkan, F.; Arslan, E.; Tütüncü, N.; Sari, A. Analysis of skin-graft loss due to infection: Infection-related graft loss. Ann. Plast. Surg. 2005, 55, 102–106. [Google Scholar] [PubMed]
- Alawi, S.A.; Limbourg, A.; Strauss, S.; Vogt, P.M. Visualisierung von Bakterien auf Verbrennungswunden und Spalthauttransplantaten nach der MEEK/MESH-Technik—Eine Pilotstudie mit ersten Erfahrungen der klinischen Wundbeurteilung durch Autofluoreszenz. Handchir. Mikrochir. Plast. Chir 2018, 50, 9. [Google Scholar] [CrossRef] [PubMed]
- Rennie, M.Y.; Jones, L.M.; Dunham, D.; Smith, A.C.; DaCosta, R.S. In vivo and in vitro detection of porphyrin-producing wound pathogens with real-time bacterial fluorescence imaging. In Proceedings of the Symposium on Advanced Wound Care (SAWC) Spring, San Antonio, TX, USA, 7–11 May 2019. [Google Scholar]
- Blomfield, J.; Farrar, J.F. The fluorescent properties of maturing arterial elastin. Cardiovasc. Res. 1969, 3, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, D. Isolation and characterization of a fluorescent material in bovine achilles tendon collagen. Biochem. Biophys. Res. Commun. 1977, 76, 1124–1129. [Google Scholar] [CrossRef]
- Zhao, H.L.; Zhang, C.P.; Zhu, H.; Jiang, Y.F.; Fu, X.B. Autofluorescence of collagen fibres in scar. Skin Res. Technol. 2017, 23, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Grey, J.E.; Enoch, S.; Harding, K.G. ABC of Wound Healing; BMJ/Blackwell: Malden, MA, USA, 2006. [Google Scholar]
- Anderson, R.R.; Parrish, J.A. The optics of human skin. J. Invest. Dermatol. 1981, 77, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, K.P.; Elsner, P.; Berardesca, E.; Maibach, H.I. Bioengineering of the Skin: Skin Imaging and Analysis, 2nd ed.; Taylor & Francis: Abingdon, UK, 1996. [Google Scholar]
- Clement, M.; Daniel, G.; Trelles, M. Optimising the design of a broad-band light source for the treatment of skin. J. Cosmet. Laser Ther. 2005, 7, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, R37–R61. [Google Scholar] [CrossRef] [PubMed]
- Petty, H.R. Fluorescence microscopy: Established and emerging methods, experimental strategies, and applications in immunology. Microsc. Res. Tech. 2007, 70, 687–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Bacteria | Tissue Components | Confounders | |
|---|---|---|---|
| Red | Porphyrin producing bacteria 1 | – |
|
| Cyan | Pseudomonas aeruginosa | – |
|
| White | Pseudomonas aeruginosa | Tendon, bone, flaky skin, nails |
|
| Dark (black/maroon) | – | Hemoglobin (blood or highly vascular tissue) Necrotic tissue |
|
| Green | – | Skin, slough |
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rennie, M.Y.; Dunham, D.; Lindvere-Teene, L.; Raizman, R.; Hill, R.; Linden, R. Understanding Real-Time Fluorescence Signals from Bacteria and Wound Tissues Observed with the MolecuLight i:XTM. Diagnostics 2019, 9, 22. https://doi.org/10.3390/diagnostics9010022
Rennie MY, Dunham D, Lindvere-Teene L, Raizman R, Hill R, Linden R. Understanding Real-Time Fluorescence Signals from Bacteria and Wound Tissues Observed with the MolecuLight i:XTM. Diagnostics. 2019; 9(1):22. https://doi.org/10.3390/diagnostics9010022
Chicago/Turabian StyleRennie, Monique Y., Danielle Dunham, Liis Lindvere-Teene, Rose Raizman, Rosemary Hill, and Ron Linden. 2019. "Understanding Real-Time Fluorescence Signals from Bacteria and Wound Tissues Observed with the MolecuLight i:XTM" Diagnostics 9, no. 1: 22. https://doi.org/10.3390/diagnostics9010022