Expression of ATP/GTP Binding Protein 1 Has Prognostic Value for the Clinical Outcomes in Non-Small Cell Lung Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Culture Condition
2.2. AGTPBP1 Knockdown Using Lentiviral Vector
2.3. Isolation of Total RNA Extraction and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.4. Cell Proliferation and Viability Assay
2.5. Cell Migration Assay
2.6. Sphere-Forming Assay
2.7. Drug Resistance Assay
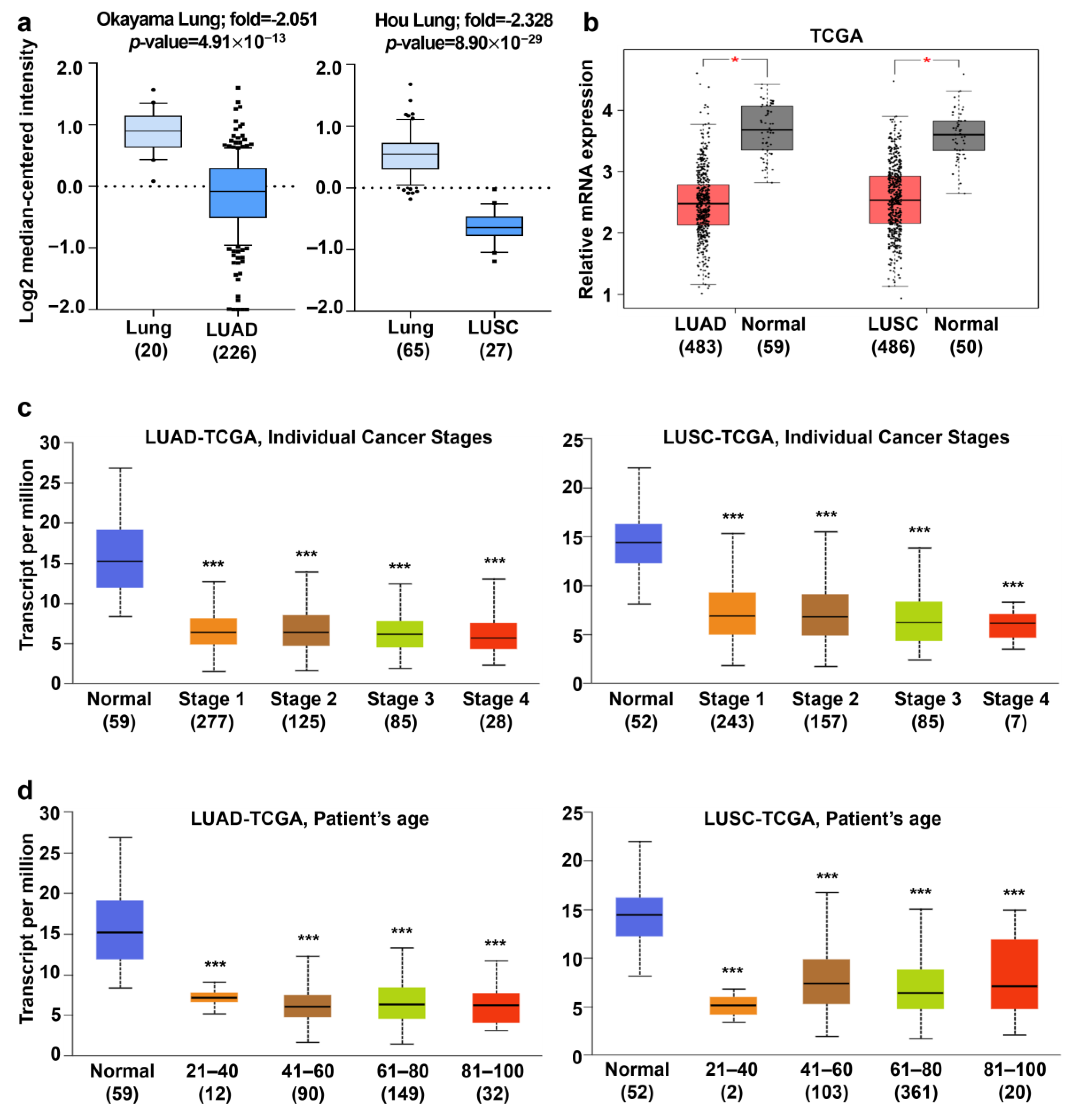
2.8. Analysis of AGTPBP1 mRNA Expression Pattern in Lung Cancer
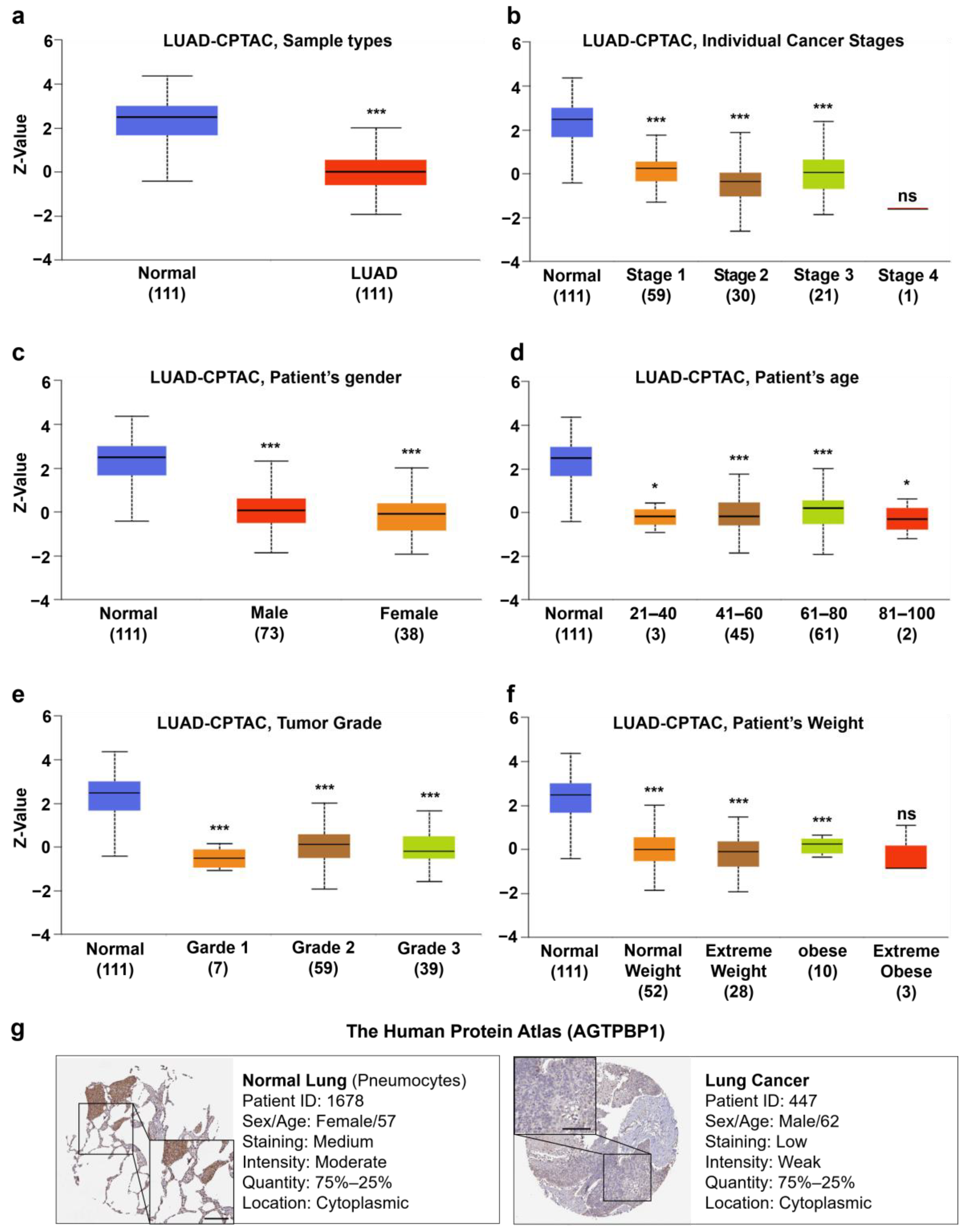
2.9. Analysis of AGTPBP1 Protein Expression Pattern in Lung Cancer
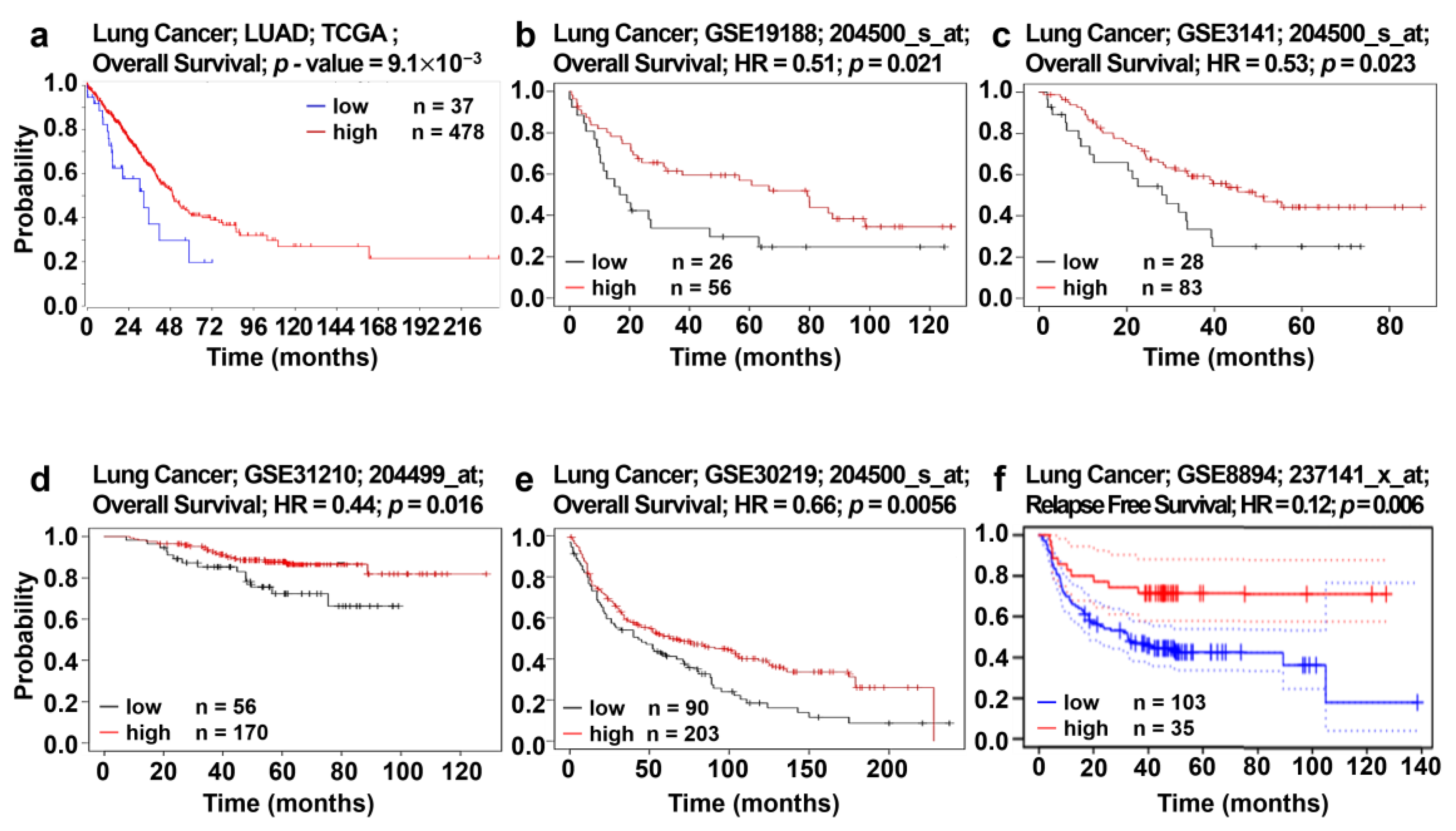
2.10. Analysis of AGTPBP1 Expression and Survival Pattern in Lung Cancer
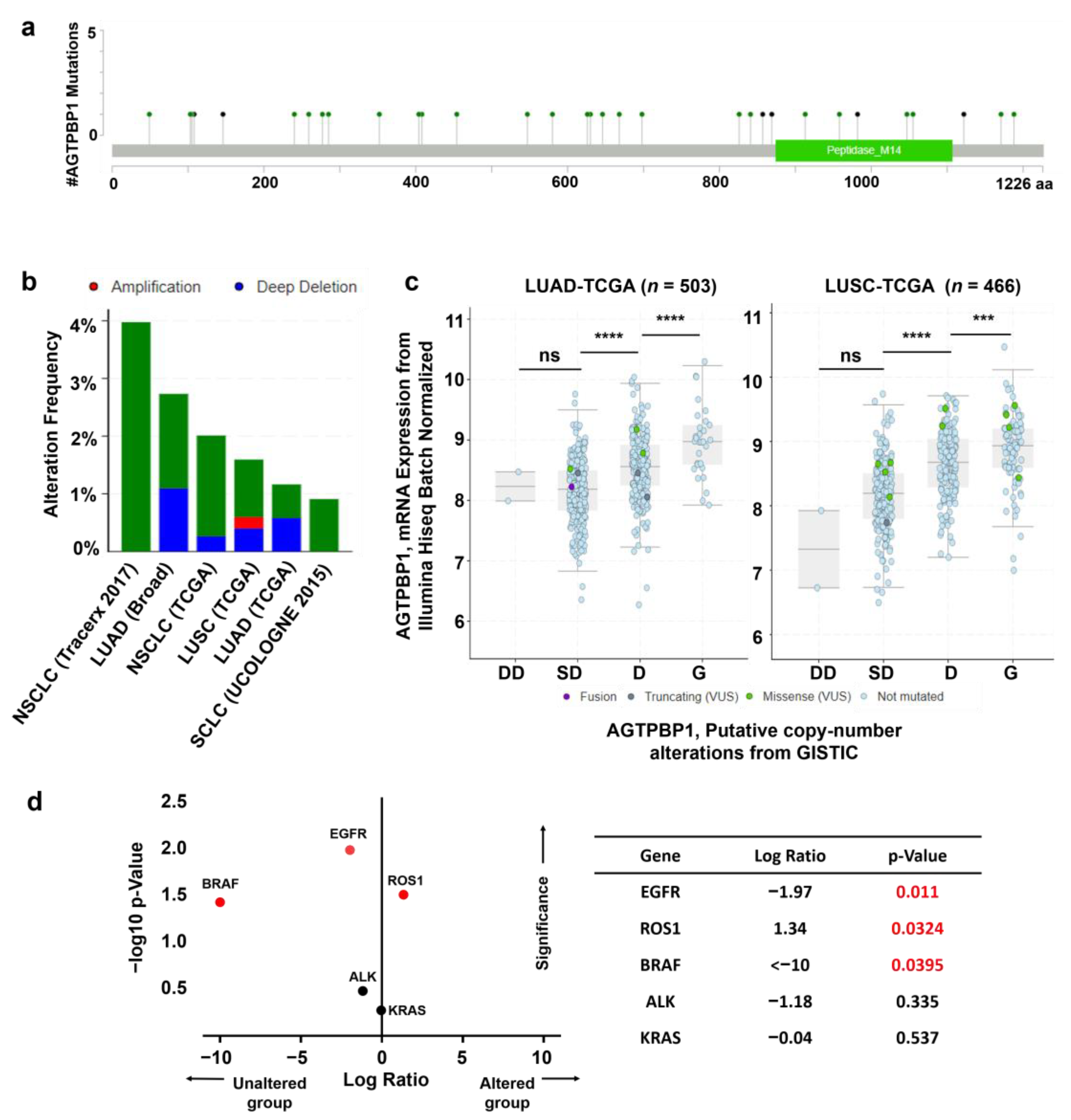
2.11. Analysis of AGTPBP1 Alteration Frequency in Lung Cancer
2.12. Analysis of Genes Co-Expressed with AGTPBP1 and Their Pathways
2.13. Analysis of Correlation between AGTPBP1 and Infiltration of Immune Cells
2.14. Statistical Analysis
3. Results
3.1. Cell Proliferation, Migration, Sphere Formation, and Drug Resistance in A549 Cells
3.2. Analysis of AGTPBP1 mRNA Expression Pattern in Lung Cancer
3.3. Analysis of AGTPBP1 Protein Level in Lung Cancer
3.4. Analysis of Mutation and Copy Number Alterations of AGTPBP1 in Lung Cancer
3.5. Analysis of Correlation between AGTPBP1 Expression and Patient Survival
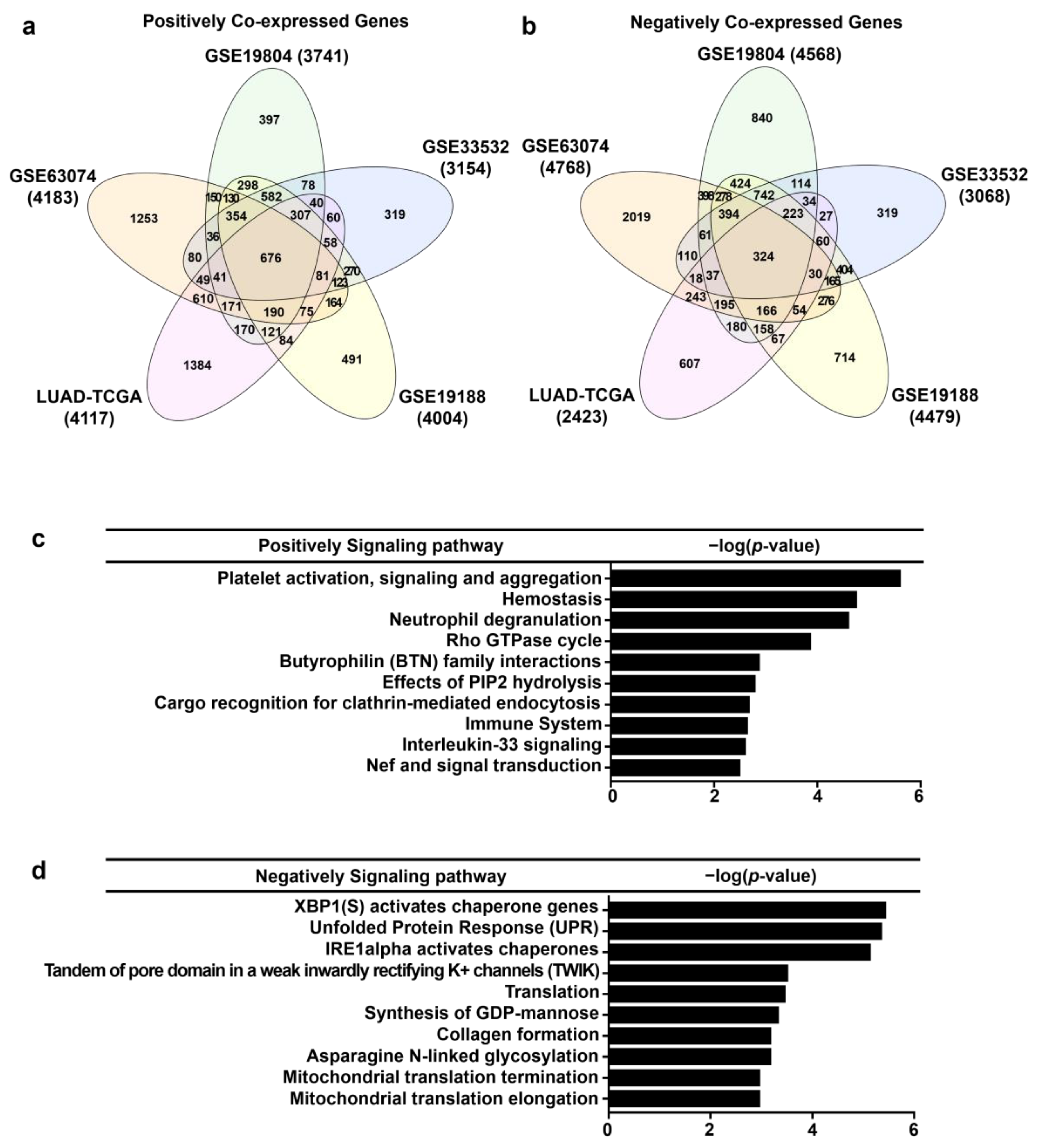
3.6. Analysis of Genes Co-Expressed with AGTPBP1 in Lung Cancer
3.7. Correlation Anlaysis between Immune Infiltration and AGTPBP1 Expression in LUAD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rogowski, K.; Van Dijk, J.; Magiera, M.M.; Bosc, C.; Deloulme, J.-C.; Bosson, A.; Peris, L.; Gold, N.D.; Lacroix, B.; Grau, M.B. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 2010, 143, 564–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, A.; Morgan, J.I.; Pecot, M.; Soumare, A.; Osborne, A.; Soares, H.D. Regenerating motor neurons express Nna1, a novel ATP/GTP-binding protein related to zinc carboxypeptidases. Mol. Cell. Neurosci. 2000, 16, 578–596. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gonzalez, A.; La Spada, A.R.; Treadaway, J.; Higdon, J.C.; Harris, B.S.; Sidman, R.L.; Morgan, J.I.; Zuo, J. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science 2002, 295, 1904–1906. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Xiao, R.; Choi, H.; Jo, H.; Kim, J.-H.; Uhm, S.-J.; Park, C. Abnormal sperm development in pcd 3J-/-mice: The importance of Agtpbp1 in spermatogenesis. Mol. Cells 2011, 31, 39–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmore-Hall, S.; Kuo, J.; Ward, J.M.; Zahra, R.; Morrison, R.S.; Perkins, G.; La Spada, A.R. CCP1 promotes mitochondrial fusion and motility to prevent Purkinje cell neuron loss in pcd mice. J. Cell Biol. 2019, 218, 206–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shashi, V.; Magiera, M.M.; Klein, D.; Zaki, M.; Schoch, K.; Rudnik-Schöneborn, S.; Norman, A.; Neto, O.L.A.; Dusl, M.; Yuan, X. Loss of tubulin deglutamylase CCP1 causes infantile-onset neurodegeneration. EMBO J. 2018, 37, e100540. [Google Scholar] [CrossRef]
- Berezniuk, I.; Vu, H.T.; Lyons, P.J.; Sironi, J.J.; Xiao, H.; Burd, B.; Setou, M.; Angeletti, R.H.; Ikegami, K.; Fricker, L.D. Cytosolic carboxypeptidase 1 is involved in processing α-and β-tubulin. J. Biol. Chem. 2012, 287, 6503–6517. [Google Scholar] [CrossRef] [Green Version]
- Audebert, S.; Desbruyeres, E.; Gruszczynski, C.; Koulakoff, A.; Gros, F.; Denoulet, P.; Eddé, B. Reversible polyglutamylation of alpha-and beta-tubulin and microtubule dynamics in mouse brain neurons. Mol. Biol. Cell 1993, 4, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Bré, M.H.; de Néchaud, B.; Wolff, A.; Fleury, A. Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell Motil. Cytoskelet. 1994, 27, 337–349. [Google Scholar] [CrossRef]
- Lessard, D.V.; Zinder, O.J.; Hotta, T.; Verhey, K.J.; Ohi, R.; Berger, C.L. Polyglutamylation of tubulin’s C-terminal tail controls pausing and motility of kinesin-3 family member KIF1A. J. Biol. Chem. 2019, 294, 6353–6363. [Google Scholar] [CrossRef]
- Kubo, T.; Yanagisawa, H.-A.; Yagi, T.; Hirono, M.; Kamiya, R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 2010, 20, 441–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobinnec, Y.; Moudjou, M.; Fouquet, J.; Desbruyeres, E.; Eddé, B.; Bornens, M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskelet. 1998, 39, 223–232. [Google Scholar] [CrossRef]
- Wloga, D.; Gaertig, J. Post-translational modifications of microtubules. J. Cell Sci. 2010, 123, 3447–3455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, V.; Kanakkanthara, A.; Chan, A.; Miller, J.H. Potential role of tubulin tyrosine ligase-like enzymes in tumorigenesis and cancer cell resistance. Cancer Lett. 2014, 350, 1–4. [Google Scholar] [CrossRef]
- Souček, K.; Kamaid, A.; Phung, A.D.; Kubala, L.; Bulinski, J.C.; Harper, R.W.; Eiserich, J.P. Normal and prostate cancer cells display distinct molecular profiles of α-tubulin posttranslational modifications. Prostate 2006, 66, 954–965. [Google Scholar] [CrossRef]
- Kato, C.; Miyazaki, K.; Nakagawa, A.; Ohira, M.; Nakamura, Y.; Ozaki, T.; Imai, T.; Nakagawara, A. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int. J. Cancer 2004, 112, 365–375. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Cucchiarelli, V.; Hiser, L.; Smith, H.; Frankfurter, A.; Spano, A.; Correia, J.J.; Lobert, S. Beta-tubulin isotype classes II and V expression patterns in nonsmall cell lung carcinomas. Cell Motil. Cytoskelet. 2008, 65, 675–685. [Google Scholar] [CrossRef]
- Gan, P.P.; McCarroll, J.A.; Po’uha, S.T.; Kamath, K.; Jordan, M.A.; Kavallaris, M. Microtubule dynamics, mitotic arrest, and apoptosis: Drug-induced differential effects of βIII-tubulin. Mol. Cancer Ther. 2010, 9, 1339–1348. [Google Scholar] [CrossRef] [Green Version]
- Christoph, D.C.; Kasper, S.; Gauler, T.C.; Loesch, C.; Engelhard, M.; Theegarten, D.; Poettgen, C.; Hepp, R.; Peglow, A.; Loewendick, H.; et al. betaV-tubulin expression is associated with outcome following taxane-based chemotherapy in non-small cell lung cancer. Br. J. Cancer 2012, 107, 823–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000135049-AGTPBP1/tissue (accessed on 3 June 2020).
- Parker, A.L.; Kavallaris, M.; McCarroll, J.A. Microtubules and their role in cellular stress in cancer. Front. Oncol. 2014, 4, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.K.; Choi, H.Y.; Yang, G.-M.; Biswas, P.K.; Kim, K.; Kang, G.-H.; Gil, M.; Cho, S.-G. GPR50 promotes hepatocellular carcinoma progression via the Notch signaling pathway through direct interaction with ADAM17. Mol. Ther. Oncolytics 2020, 17, 332–349. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 1–9. [Google Scholar]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Koster, J.; Volckmann, R.; Zwijnenburg, D.; Molenaar, P.; Versteeg, R. R2: Genomics Analysis and Visualization Platform. Cancer Res. 2019, 79 (Suppl. 13), 2490. [Google Scholar]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G. Impact of genetic polymorphisms on human immune cell gene expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Aerts, J.; Den Hamer, B.; Van Ijcken, W.; Den Bakker, M.; Riegman, P.; van der Leest, C.; van der Spek, P.; Foekens, J.A.; Hoogsteden, H.C. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef] [PubMed]
- Bild, A.H.; Yao, G.; Chang, J.T.; Wang, Q.; Potti, A.; Chasse, D.; Joshi, M.-B.; Harpole, D.; Lancaster, J.M.; Berchuck, A. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006, 439, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, S.; Debernardi, A.; Jacquiau, B.; Vitte, A.-L.; Vesin, A.; Nagy-Mignotte, H.; Moro-Sibilot, D.; Brichon, P.-Y.; Lantuejoul, S.; Hainaut, P. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci. Transl. Med. 2013, 5, ra166–ra186. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.-S.; Son, D.-S.; Kim, S.-H.; Lee, J.; Jo, J.; Han, J.; Kim, H.; Lee, H.J.; Choi, H.Y.; Jung, Y. Prediction of recurrence-free survival in postoperative non–small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin. Cancer Res. 2008, 14, 7397–7404. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Reitze, N.J.; Ewing, A.L.; McCreary, S.; Uihlein, A.H.; Brower, S.L.; Wang, D.; Wang, T.; Gabrin, M.J.; Keating, K.E. Analytical performance of a 15-gene prognostic assay for early-stage non–small-cell lung carcinoma using RNA-stabilized tissue. J. Mol. Diagn. 2015, 17, 438–445. [Google Scholar] [CrossRef]
- Lu, T.-P.; Tsai, M.-H.; Lee, J.-M.; Hsu, C.-P.; Chen, P.-C.; Lin, C.-W.; Shih, J.-Y.; Yang, P.-C.; Hsiao, C.K.; Lai, L.-C. Identification of a novel biomarker, sema5a, for non–small cell lung carcinoma in nonsmoking women. Cancer Epidemiol. Prev. Biomark. 2010, 19, 2590–2597. [Google Scholar] [CrossRef] [Green Version]
- Meister, M.; Belousov, A.; Xu, E.; Schnabel, P.; Warth, A.; Hoofmann, H. Intra-tumor heterogeneity of gene expression profiles in early stage non-small cell lung cancer. J. Bioinf. Res. Stud. 2014, 1, 1. [Google Scholar]
- Vivier, E.; Nunès, J.A.; Vély, F. Natural killer cell signaling pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Hayakawa, Y.; Smyth, M.J.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Karunakaran, M.M.; Willcox, C.R.; Salim, M.; Paletta, D.; Fichtner, A.S.; Noll, A.; Starick, L.; Nöhren, A.; Begley, C.R.; Berwick, K.A. Butyrophilin-2A1 directly binds germline-encoded regions of the Vγ9Vδ2 TCR and is essential for phosphoantigen sensing. Immunity 2020, 52, 487–498.e6. [Google Scholar] [CrossRef]
- Chew, V.; Toh, H.C.; Abastado, J.-P. Immune microenvironment in tumor progression: Characteristics and challenges for therapy. J. Oncol. 2012, 2012, 608406. [Google Scholar] [CrossRef] [Green Version]
- Eruslanov, E.B.; Bhojnagarwala, P.S.; Quatromoni, J.G.; Stephen, T.L.; Ranganathan, A.; Deshpande, C.; Akimova, T.; Vachani, A.; Litzky, L.; Hancock, W.W. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Investig. 2014, 124, 5466–5480. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Feng, Y.; Yue, C.; Xu, B.; Chen, L.; Jiang, J.; Lu, B.; Zhu, Y. Lower expression level of IL-33 is associated with poor prognosis of pulmonary adenocarcinoma. PLoS ONE 2018, 13, e0193428. [Google Scholar] [CrossRef]
- Bumpers, H.L.; Huang, M.-B.; Powell, M.; Grizzle, W.E.; Lillard, J.; Okoli, J.; Bond, V.C. Effects of HIV-1 Nef, a cytotoxic viral protein, on the growth of primary colorectal cancer. Cancer Biol. Ther. 2005, 4, 72–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiler, C.; Gebhart, N.; Zhang, Y.; Shinton, S.A.; Li, Y.-S.; Ross, N.L.; Liu, X.; Li, Q.; Bilbee, A.N.; Varshney, G.K. Mutagenesis Screen Identifies agtpbp1 and eps15L1 as Essential for T lymphocyte Development in Zebrafish. PLoS ONE 2015, 10, e0131908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Hu, Y.; Yao, C. The paradoxical role of tumor-infiltrating immune cells in lung cancer. Intractable Rare Dis. Res. 2017, 6, 234–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakimi, K.; Matsushita, H.; Murakawa, T.; Nakajima, J. γδ T cell therapy for the treatment of non-small cell lung cancer. Transl. Lung Cancer Res. 2014, 3, 23. [Google Scholar]
- Kyuhou, S.-i.; Kato, N.; Gemba, H. Emergence of endoplasmic reticulum stress and activated microglia in Purkinje cell degeneration mice. Neurosci. Lett. 2006, 396, 91–96. [Google Scholar] [CrossRef]
- Yadav, R.K.; Chae, S.-W.; Kim, H.-R.; Chae, H.J. Endoplasmic reticulum stress and cancer. J. Cancer Prev. 2014, 19, 75. [Google Scholar] [CrossRef]
- Koong, A.C.; Chauhan, V.; Romero-Ramirez, L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol. Ther. 2006, 5, 756–759. [Google Scholar] [CrossRef] [Green Version]
- Pluquet, O.; Dejeans, N.; Bouchecareilh, M.; Lhomond, S.; Pineau, R.; Higa, A.; Delugin, M.; Combe, C.; Loriot, S.; Cubel, G. Posttranscriptional regulation of PER1 underlies the oncogenic function of IREα. Cancer Res. 2013, 73, 4732–4743. [Google Scholar] [CrossRef] [Green Version]
- Cubillos-Ruiz, J.R.; Silberman, P.C.; Rutkowski, M.R.; Chopra, S.; Perales-Puchalt, A.; Song, M.; Zhang, S.; Bettigole, S.E.; Gupta, D.; Holcomb, K. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell 2015, 161, 1527–1538. [Google Scholar] [CrossRef] [Green Version]
- Kalinina, E.; Biswas, R.; Berezniuk, I.; Hermoso, A.; Aviles, F.X.; Flicker, L.D. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 2007, 21, 836–850. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef] [PubMed]
- McCarroll, J.A.; Gan, P.P.; Liu, M.; Kavallaris, M. βIII-Tubulin Is a Multifunctional Protein Involved in Drug Sensitivity and Tumorigenesis in Non–Small Cell Lung Cancer. Cancer Res. 2010, 70, 4995–5003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxevanos, P.; Mountzios, G. Novel chemotherapy regimens for advanced lung cancer: Have we reached a plateau? Ann. Transl. Med. 2018, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Cao, L.; Zhu, C.; Xie, H.; Hao, K.; Xia, N.; Wang, B.; Zhang, Y.; Liu, F. Improving lung cancer treatment: Hyaluronic acidmodified and glutathioneresponsive amphiphilic TPGSdoxorubicin prodrugentrapped nanoparticles. Oncol. Rep. 2019, 42, 361–369. [Google Scholar] [CrossRef]
- Cabeza, L.; Ortiz, R.; Prados, J.; Delgado, A.V.; Martin-Villena, M.J.; Clares, B.; Perazzoli, G.; Entrena, J.M.; Melguizo, C.; Arias, J.L. Improved antitumor activity and reduced toxicity of doxorubicin encapsulated in poly(epsilon-caprolactone) nanoparticles in lung and breast cancer treatment: An in vitro and in vivo study. Eur. J. Pharm. Sci. 2017, 102, 24–34. [Google Scholar] [CrossRef]
- Hong, Y.; Che, S.; Hui, B.; Yang, Y.; Wang, X.; Zhang, X.; Qiang, Y.; Ma, H. Lung cancer therapy using doxorubicin and curcumin combination: Targeted prodrug based, pH sensitive nanomedicine. Biomed. Pharm. 2019, 112, 108614. [Google Scholar] [CrossRef]
- Cao, C.; Wang, Q.; Liu, Y. Lung cancer combination therapy: Doxorubicin and beta-elemene co-loaded, pH-sensitive nanostructured lipid carriers. Drug Des. Dev. Ther. 2019, 13, 1087–1098. [Google Scholar] [CrossRef] [Green Version]
- Poornima, P.; Kumar, V.B.; Weng, C.F.; Padma, V.V. Doxorubicin induced apoptosis was potentiated by neferine in human lung adenocarcima, A549 cells. Food Chem. Toxicol. 2014, 68, 87–98. [Google Scholar] [CrossRef]
- Guo, J.; Xu, B.; Han, Q.; Zhou, H.; Xia, Y.; Gong, C.; Dai, X.; Li, Z.; Wu, G. Ferroptosis: A Novel Anti-tumor Action for Cisplatin. Cancer Res. Treat. 2018, 50, 445–460. [Google Scholar] [CrossRef] [Green Version]
- Calvayrac, O.; Pradines, A.; Pons, E.; Mazières, J.; Guibert, N. Molecular biomarkers for lung adenocarcinoma. Eur. Respir. J. 2017, 49, 1601734. [Google Scholar] [CrossRef]
- Gkolfinopoulos, S.; Mountzios, G. Beyond EGFR and ALK: Targeting rare mutations in advanced non-small cell lung cancer. Ann. Transl. Med. 2018, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qu, J.; Xu, Z. Clinical features and mutation status of EGFR, KRAS, BRAF, EML4-ALK and ROS1 between surgical resection samples and non surgical resection samples in lung cancer. J. Thorac. Dis. 2015, 7, 875. [Google Scholar] [PubMed]
- Yoon, Y.K.; Kim, H.P.; Han, S.W.; Oh, D.Y.; Im, S.A.; Bang, Y.J.; Kim, T.Y. KRAS mutant lung cancer cells are differentially responsive to MEK inhibitor due to AKT or STAT3 activation: Implication for combinatorial approach. Mol. Carcinog. 2010, 49, 353–362. [Google Scholar]
- Pao, W.; Wang, T.Y.; Riely, G.J.; Miller, V.A.; Pan, Q.; Ladanyi, M.; Zakowski, M.F.; Heelan, R.T.; Kris, M.G.; Varmus, H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005, 2, e17. [Google Scholar] [CrossRef] [Green Version]
- Rodenhuis, S.; Slebos, R.J.; Boot, A.J.; Evers, S.G.; Mooi, W.J.; Wagenaar, S.S.; van Bodegom, P.C.; Bos, J.L. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988, 48, 5738–5741. [Google Scholar]
- Suzuki, Y.; Orita, M.; Shiraishi, M.; Hayashi, K.; Sekiya, T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene 1990, 5, 1037–1043. [Google Scholar]
- Bergethon, K.; Shaw, A.T.; Ou, S.-H.I.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863. [Google Scholar] [CrossRef] [Green Version]
- Hiraki, S.M.; Seto, T.; Tamura, T.; Watanabe, Y.; Ozawa, S.; Ikeda, H.; Nakata, Y.; Ohnoshi, T.; Kimura, I. Establishment of human continuous cell lines from squamous cell, adeno-and small cell carcinoma of the lung and the results of hetero-transplantation. Haigan 1982, 22, 53–58. [Google Scholar] [CrossRef]


| Description | LUAD | LUSC | ||
|---|---|---|---|---|
| Cor | p | Cor | p | |
| Purity | −0.119 | * | 0.069 | 0.133 |
| B Cell | 0.204 | *** | 0.135 | * |
| CD8+ T Cell | 0.257 | *** | 0.144 | * |
| CD4+ T Cell | 0.295 | *** | 0.132 | * |
| Macrophage | 0.298 | *** | 0.063 | 0.169 |
| Neutrophil | 0.359 | *** | 0.072 | 0.114 |
| Dendritic Cell | 0.271 | *** | 0.141 | * |
| Description | Gene Marker | LUAD | LUSC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | ||||||
| Cor | p | Cor | p | Cor | p | Cor | p | ||
| CD8+ T cell | CD8A | 0.332 | *** | 0.339 | *** | 0.161 | ** | 0.158 | ** |
| CD8B | 0.231 | *** | 0.233 | *** | 0.126 | * | 0.113 | 0.014 | |
| T cell (general) | CD3D | 0.180 | *** | 0.185 | *** | 0.092 | 0.040 | 0.090 | 0.050 |
| CD3E | 0.293 | *** | 0.305 | *** | 0.127 | * | 0.127 | * | |
| CD2 | 0.260 | *** | 0.269 | *** | 0.128 | * | 0.121 | * | |
| B cell | CD19 | 0.184 | *** | 0.199 | *** | 0.106 | 0.018 | 0.110 | 0.017 |
| CD79A | 0.131 | * | 0.147 | * | 0.089 | 0.046 | 0.094 | 0.040 | |
| Monocyte | CD86 | 0.331 | *** | 0.331 | *** | 0.154 | ** | 0.143 | * |
| CD115 (CSF1R) | 0.303 | *** | 0.303 | *** | 0.150 | ** | 0.139 | * | |
| TAM | CCL2 | 0.179 | *** | 0.186 | *** | 0.066 | 0.141 | 0.063 | 0.168 |
| CD68 | 0.252 | *** | 0.261 | *** | 0.088 | 0.048 | 0.087 | 0.058 | |
| IL10 | 0.278 | *** | 0.280 | *** | −0.002 | 0.965 | 0.003 | 0.947 | |
| M1 Macrophage | NOS2 | 0.262 | *** | 0.262 | *** | 0.108 | 0.016 | 0.119 | * |
| IRF5 | 0.267 | *** | 0.277 | *** | 0.070 | 0.117 | 0.060 | 0.188 | |
| COX2 (PTGS2) | 0.010 | 0.825 | 0.007 | 0.880 | −0.075 | 0.095 | −0.074 | 0.107 | |
| M2 Macrophage | CD163 | 0.408 | *** | 0.408 | *** | 0.142 | * | 0.134 | * |
| VSIG4 | 0.284 | *** | 0.285 | *** | 0.066 | 0.139 | 0.056 | 0.219 | |
| MS4A4A | 0.322 | *** | 0.327 | *** | 0.114 | 0.010 | 0.105 | 0.022 | |
| Neutrophil | CEACAM8 | 0.117 | * | 0.127 | * | −0.021 | 0.642 | −0.023 | 0.615 |
| ITGAM | 0.306 | *** | 0.316 | *** | 0.190 | *** | 0.179 | *** | |
| CCR7 | 0.263 | *** | 0.275 | *** | 0.169 | ** | 0.171 | ** | |
| NK cell | KIR2DL1 | 0.206 | *** | 0.210 | *** | 0.038 | 0.392 | 0.052 | 0.256 |
| KIR2DL3 | 0.235 | *** | 0.240 | *** | 0.054 | 0.231 | 0.035 | 0.445 | |
| KIR2DL4 | 0.178 | *** | 0.178 | *** | 0.034 | 0.446 | 0.022 | 0.634 | |
| KIR3DL1 | 0.208 | *** | 0.196 | *** | 0.089 | 0.046 | 0.077 | 0.093 | |
| KIR3DL2 | 0.206 | *** | 0.209 | *** | 0.107 | 0.017 | 0.109 | 0.017 | |
| KIR3DL3 | 0.159 | ** | 0.156 | ** | 0.064 | 0.152 | 0.075 | 0.103 | |
| KIR2DS4 | 0.216 | *** | 0.218 | *** | 0.056 | 0.209 | 0.072 | 0.115 | |
| Dendritic cell | HLA-DPB1 | 0.155 | ** | 0.161 | ** | 0.122 | * | 0.112 | 0.014 |
| HLA-DQB1 | 0.078 | 0.078 | 0.083 | 0.066 | 0.062 | 0.164 | 0.055 | 0.229 | |
| HLA-DRA | 0.139 | * | 0.143 | * | 0.098 | 0.028 | 0.085 | 0.064 | |
| HLA-DPA1 | 0.181 | *** | 0.186 | *** | 0.136 | * | 0.124 | * | |
| BDCA-1 (CD1C) | 0.066 | 0.133 | 0.070 | 0.121 | 0.046 | 0.305 | 0.041 | 0.367 | |
| BDCA-4 (NRP1) | 0.223 | *** | 0.217 | *** | 0.062 | 0.164 | 0.061 | 0.180 | |
| CD11c (ITGAX) | 0.377 | *** | 0.383 | *** | 0.176 | *** | 0.175 | ** | |
| Th1 | T-bet (TBX21) | 0.350 | *** | 0.362 | *** | 0.205 | *** | 0.206 | *** |
| STAT4 | 0.188 | *** | 0.200 | *** | 0.065 | 0.145 | 0.063 | 0.172 | |
| STAT1 | 0.339 | *** | 0.347 | *** | 0.139 | * | 0.141 | * | |
| IFN-γ (IFNG) | 0.236 | *** | 0.247 | *** | 0.099 | 0.027 | 0.098 | 0.033 | |
| TNF-α (TNF) | 0.143 | * | 0.145 | * | −0.044 | 0.322 | −0.052 | 0.258 | |
| Th2 | GATA3 | 0.261 | *** | 0.273 | *** | −0.125 | * | −0.136 | * |
| STAT6 | 0.151 | ** | 0.167 | ** | 0.074 | 0.099 | 0.074 | 0.106 | |
| STAT5A | 0.429 | *** | 0.438 | *** | 0.239 | *** | 0.237 | *** | |
| IL13 | 0.088 | 0.046 | 0.104 | 0.021 | 0.202 | *** | 0.202 | *** | |
| Tfh | BCL6 | 0.204 | *** | 0.208 | *** | 0.223 | *** | 0.233 | *** |
| IL21 | 0.211 | *** | 0.218 | *** | 0.113 | 0.011 | 0.104 | 0.022 | |
| Th17 | STAT3 | 0.251 | *** | 0.249 | *** | 0.208 | *** | 0.215 | *** |
| IL17A | 0.101 | 0.022 | 0.108 | 0.017 | 0.035 | 0.440 | 0.031 | 0.493 | |
| Treg | FOXP3 | 0.222 | *** | 0.232 | *** | 0.154 | ** | 0.146 | * |
| CCR8 | 0.315 | *** | 0.326 | *** | 0.200 | *** | 0.193 | *** | |
| STAT5B | 0.478 | *** | 0.483 | *** | 0.355 | *** | 0.360 | *** | |
| TGFβ (TGFB1) | 0.177 | *** | 0.177 | *** | −0.161 | ** | −0.148 | * | |
| Exhausted T cell | PD-1 (PDCD1) | 0.235 | *** | 0.237 | *** | 0.154 | ** | 0.154 | ** |
| CTLA4 | 0.290 | *** | 0.301 | *** | 0.168 | ** | 0.164 | ** | |
| LAG3 | 0.241 | *** | 0.248 | *** | 0.130 | * | 0.122 | * | |
| TIM-3 (HAVCR2) | 0.311 | *** | 0.313 | *** | 0.141 | * | 0.128 | * | |
| Description | Gene Marker | LUAD | LUSC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | ||||||
| Cor | p | Cor | p | Cor | p | Cor | p | ||
| Activation receptors | KLRK1 | 0.350 | *** | 0.358 | *** | 0.183 | *** | 0.182 | *** |
| NCR1 | 0.306 | *** | 0.309 | *** | 0.182 | *** | 0.168 | ** | |
| NCR2 | 0.045 | 0.312 | 0.066 | 0.145 | 0.029 | 0.518 | 0.036 | 0.435 | |
| NCR3 | 0.228 | *** | 0.242 | *** | 0.124 | * | 0.131 | * | |
| FAS/FASL | FAS | 0.181 | *** | 0.190 | *** | 0.052 | 0.247 | 0.064 | 0.163 |
| FASLG | 0.284 | *** | 0.291 | *** | 0.108 | 0.016 | 0.106 | 0.020 | |
| Cytolytic molecules | GZMA | 0.255 | *** | 0.257 | *** | 0.053 | 0.238 | 0.050 | 0.271 |
| GZMB | 0.252 | *** | 0.256 | *** | 0.105 | 0.019 | 0.106 | 0.020 | |
| PRF1 | 0.217 | *** | 0.221 | *** | 0.126 | * | 0.127 | * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwak, H.J.; Gil, M.; Chae, H.S.; Seok, J.; Soundrarajan, N.; Saha, S.K.; Kim, A.; Park, K.S.; Park, C.; Cho, S.-G. Expression of ATP/GTP Binding Protein 1 Has Prognostic Value for the Clinical Outcomes in Non-Small Cell Lung Carcinoma. J. Pers. Med. 2020, 10, 263. https://doi.org/10.3390/jpm10040263
Kwak HJ, Gil M, Chae HS, Seok J, Soundrarajan N, Saha SK, Kim A, Park KS, Park C, Cho S-G. Expression of ATP/GTP Binding Protein 1 Has Prognostic Value for the Clinical Outcomes in Non-Small Cell Lung Carcinoma. Journal of Personalized Medicine. 2020; 10(4):263. https://doi.org/10.3390/jpm10040263
Chicago/Turabian StyleKwak, Hee Jeong, Minchan Gil, Hee Sung Chae, Jaekwon Seok, Nagasundarapandian Soundrarajan, Subbroto Kumar Saha, Aram Kim, Kyoung Sik Park, Chankyu Park, and Ssang-Goo Cho. 2020. "Expression of ATP/GTP Binding Protein 1 Has Prognostic Value for the Clinical Outcomes in Non-Small Cell Lung Carcinoma" Journal of Personalized Medicine 10, no. 4: 263. https://doi.org/10.3390/jpm10040263








