Genomic Association of Single Nucleotide Polymorphisms with Blood Pressure Response to Hydrochlorothiazide among South African Adults with Hypertension
Abstract
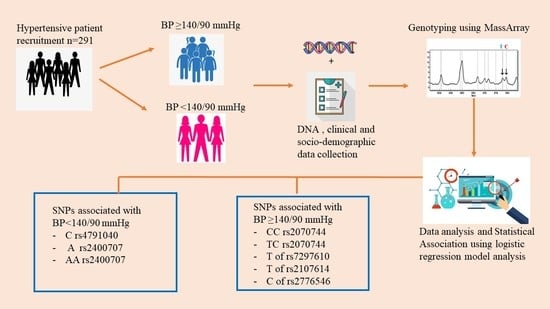
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Patient Selection
2.3. Data Collection
2.4. DNA Isolation
2.5. Genotyping
2.6. Statistical Analysis
2.7. Selection of Pharmacogenomics Biomarkers
3. Results
3.1. General Characteristics of the Study
3.2. Expression Patterns of Single Nucleotide Polymorphisms
3.3. Association between SNPs and Blood Pressure Response to Hydrochlorothiazide
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data
References
- Unger, T.; Borghi, C.; Charchar, F.; Khan, N.A.; Poulter, N.R.; Prabhakaran, D.; Ramirez, A.; Schlaich, M.; Stergiou, G.S.; Tomaszewski, M.; et al. 2020 International Society of Hypertension Global Hypertension Practice Guidelines. Hypertension 2020, 75, 1334–1357. [Google Scholar] [CrossRef]
- Heart & Stroke Foundation South Africa. Available online: https://www.heartfoundation.co.za/ (accessed on 23 September 2020).
- Jongen, V.W.; Lalla-Edward, S.T.; Vos, A.G.; Godijk, N.G.; Tempelman, H.; Grobbee, D.E.; Devillé, W.; Klipstein-Grobusch, K. Hypertension in a rural community in South Africa: What they know, what they think they know and what they recommend. BMC Public Health 2019, 19, 341. [Google Scholar] [CrossRef]
- Wagstaff, A.J. Valsartan/hydrochlorothiazide. Drugs 2006, 66, 1881–1901. [Google Scholar] [CrossRef]
- Duarte, J.D.; Cooper-DeHoff, R.M. Mechanisms for blood pressure lowering and metabolic effects of thiazide and thiazide-like diuretics. Expert Rev. Cardiovasc. 2010, 8, 793–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahin, M.H.; Gong, Y.; McDonough, C.W.; Rotroff, D.M.; Beitelshees, A.L.; Garrett, T.J.; Gums, J.G.; Motsinger-Reif, A.; Chapman, A.B.; Turner, S.T.; et al. A genetic response score for hydrochlorothiazide use: Insights from genomics and metabolomics integration. Hypertension 2016, 68, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, G.L.; Turner, S.T. Pharmacogenetics of antihypertensive drug responses. Am. J. Pharm. 2004, 1, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.T.; Schwartz, G.L.; Chapman, A.B.; Boerwinkle, E. WNK1 kinase polymorphism and blood pressure response to a thiazide diuretic. Hypertension 2005, 46, 758–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamil, S.; Erdmann, J.; Abdalrahman, I.B.; Mohamed, A.O. Association of NOS3 gene polymorphisms with essential hypertension in Sudanese patients: A case control study. BMC Med. Genet. 2017, 18, 128. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Paula, G.H.; Luizon, M.R.; Lacchini, R.; Fontana, V.; Silva, P.S.; Biagi, C.; Tanus-Santos, J.E. Gene–gene interactions among PRKCA, NOS3 and BDKRB2 polymorphisms affect the antihypertensive effects of enalapril. Basic Clin. Pharmacol. Toxicol. 2017, 120, 284–291. [Google Scholar] [CrossRef] [Green Version]
- Braz, J.C.; Gregory, K.; Pathak, A.; Zhao, W.; Sahin, B.; Klevitsky, R.; Kimball, T.F.; Lorenz, J.N.; Nairn, A.C.; Liggett, S.B.; et al. PKC-α regulates cardiac contractility and propensity toward heart failure. Nat. Med. 2004, 10, 248–254. [Google Scholar] [CrossRef]
- Bergaya, S.; Faure, S.; Baudrie, V.; Rio, M.; Escoubet, B.; Bonnin, P.; Henrion, D.; Loirand, G.; Achard, J.M.; Jeunemaitre, X.; et al. WNK1 Regulates Vasoconstriction and Blood Pressure Response to α1-Adrenergic Stimulation in Mice. Hypertension 2011, 58, 439–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoorn, E.J.; Ellison, D.H. WNK kinases and the kidney. Exp. Cell Res. 2012, 318, 1020–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S.T.; Boerwinkle, E.; O’Connell, J.R.; Bailey, K.R.; Gong, Y.; Chapman, A.B.; McDonough, C.W.; Beitelshees, A.L.; Schwartz, G.L.; Gums, J.G.; et al. Genomic Association Analysis of Common Variants Influencing Antihypertensive Response to Hydrochlorothiazide. Hypertension 2013, 62, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kohli, S.; Mishra, A.; Garg, R.; Alam, P.; Stobdan, T.; Nejatizadeh, A.; Gupta, M.; Tyagi, S.; Pasha, M.A. Interactions Between the Genes of Vasodilatation Pathways Influence Blood Pressure and Nitric Oxide Level in Hypertension. Am. J. Hypertens. 2015, 1, 239–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, P.S.; Fontana, V.; Luizon, M.R.; Lacchini, R.; Silva, W.A.; Biagi, C.; Tanus-Santos, J.E. eNOS and BDKRB2 genotypes affect the antihypertensive responses to enalapril. Eur. J. Clin. Pharmacol. 2013, 69, 167–177. [Google Scholar] [CrossRef]
- Hsu, C.C.; Shi, J.; Yuan, C.; Zhao, D.; Jiang, S.; Lyu, J.; Wang, X.; Li, H.; Wen, H.; Li, W.; et al. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A. Z deposition in non-small cell lung cancer. Genes Dev. 2018, 32, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Duarte, J.D.; Turner, S.T.; Tran, B.; Chapman, A.B.; Bailey, K.R.; Gong, Y.; Gums, J.G.; Langaee, T.Y.; Beitelshees, A.L.; Cooper-Dehoff, R.M.; et al. Association of Chromosome 12 locus with antihypertensive response to hydrochlorothiazide may involve differential YEATS4 expression. Pharm. J. 2013, 13, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Cooper-DeHoff, R.M.; Johnson, J.A. Hypertension pharmacogenomics: In search of personalized treatment approaches. Nat. Rev. Nephrol. 2016, 12, 110. [Google Scholar] [CrossRef] [Green Version]
- Daly, A.K. Pharmacogenomics of adverse drug reactions. Genome Med. 2013, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.A. Pharmacogenomics of antihypertensive drugs: Past, present and future. Pharmacogenomics 2010, 11, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Alwi, Z.B. The use of SNPs in pharmacogenomics studies. Malays. J. Med. Sci. 2005, 12, 4. [Google Scholar] [PubMed]
- Ensembl Genome Browser 100. Available online: https://www.ensembl.org/index.html (accessed on 7 June 2020).
- Jarari, N.; Rao, N.; Peela, J.R.; Ellafi, K.A.; Shakila, S.; Said, A.R.; Nelapalli, N.K.; Min, Y.; Tun, K.D.; Jamallulail, S.I.; et al. A review on prescribing patterns of antihypertensive drugs. Clin. Hypertens. 2015, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, F.; Hirbo, J.; Tishkoff, S.A. Genetic variation and adaptation in Africa: Implications for human evolution and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a008524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, D.M.; Elliott, G.S.; Chute, H.; Horan, T.; Pfenninger, K.H.; Sanford, S.D.; Foster, S.; Scully, S.; Welcher, A.A.; Holers, V.M. CSMD1 Is a Novel Multiple Domain Complement-Regulatory Protein Highly Expressed in the Central Nervous System and Epithelial Tissues. J. Immunol. 2006, 176, 4419–4430. [Google Scholar] [CrossRef] [PubMed]
- Chittani, M.; Zaninello, R.; Lanzani, C.; Frau, F.; Ortu, M.F.; Salvi, E.; Fresu, G.; Citterio, L.; Braga, D.; Piras, D.A.; et al. TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J. Hypertens. 2015, 33, 1301–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, K.W.; Go, M.J.; Jin, H.S.; Lim, J.E.; Lee, J.Y.; Han, B.G.; Hwang, S.Y.; Lee, S.H.; Park, H.K.; Cho, Y.S.; et al. Genetic variations in ATP2B1, CSK, ARSG and CSMD1 loci are related to blood pressure and/or hypertension in two Korean cohorts. J. Hum. Hypertens. 2010, 24, 367–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PRKCA Protein Kinase C Alpha [Homo Sapiens (Human)]—Gene—NCBI. Available online: https://www.ncbi.nlm.nih.gov/gene/5578 (accessed on 24 September 2020).
- Cruz-González, I.; Corral, E.; Sánchez-Ledesma, M.; Sánchez-Rodríguez, A.; Martín-Luengo, C.; González-Sarmiento, R. Association between-T786C NOS3 polymorphism and resistant hypertension: A prospective cohort study. BMC Cardiovasc. Disord. 2009, 9, 35. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.T.; Bailey, K.R.; Fridley, B.L.; Chapman, A.B.; Schwartz, G.L.; Chai, H.S.; Sicotte, H.; Kocher, J.-P.; Rodin, A.S.; Boerwinkle, E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension 2008, 52, 359–365. [Google Scholar] [CrossRef] [Green Version]
| Variables | All Participants (n; %) | Males (n; %) | Females (n; %) |
|---|---|---|---|
| All | 291(100) | 78(26.04) | 213(73.19) |
| Age (Years) | |||
| 18–25 | 01(0.34) | 01(1.28) | 0(0.00) |
| 26–35 | 08(2.75) | 02(2.56) | 06(2.82) |
| 36–45 | 19(6.52) | 07(8.97) | 12(6.63) |
| 46–55 | 52(17.87) | 14(17.95) | 38(17.84) |
| 56–65 | 120(41.24) | 25(32.05) | 95(44.60) |
| ≥66 | 91(31.27) | 29(37.18) | 62(29.11) |
| Ethnicity | |||
| Zulu | 112(38.49) | 19(24.36) | 93(43.66) |
| Swati | 19(6.53) | 03(3.85) | 16(7.51) |
| Xhosa | 160(54.98) | 56(71.79) | 104(48.83) |
| Smoking status | |||
| Never smoked | 196(67.35) | 30(38.46) | 166(77.93) |
| Ever smoked | 95(32.65) | 48(61.54) | 47(22.07) |
| Salt intake | |||
| Low-moderate | 237(81.44) | 58(74.36) | 179(84.04) |
| Increased | 54(18.56) | 20(25.64) | 34(15.96) |
| Blood pressure | |||
| <140/90 mmHg | 91(31.26) | 16(20.51) | 75(35.21) |
| ≥140/90 mmHg | 200(68.73) | 62(79.49) | 138(64.79) |
| Drug regime | |||
| HCTZ alone | 63(21.65) | 20(25.64) | 43(20.19) |
| HCTZ + 1 drug | 127(43.64) | 26(33.33) | 101(47.42) |
| HCTZ+ 2 drugs | 98(33.68) | 30(38.46) | 68(31.92) |
| HCTZ + 3 drugs | 03(1.03) | 02(2.56) | 01(0.47) |
| dbSNP | Gene | Ethnic Groups | Gender | Age | |||||
|---|---|---|---|---|---|---|---|---|---|
| Zulu (n; %) | Swati (n; %) | Xhosa (n; %) | Male (n; %) | Female (n; %) | <55 Years | 55–65 Years | >65 Years | ||
| All | 112(38.48) | 19(6.52) | 160(54.98) | 78(26.80) | 213(73.19) | 80(27.49) | 121(41.58) | 90(30.93) | |
| rs11189015 | SLIT1 | ||||||||
| Yes | - | - | 155(96.88) | 54(69.23) | 101(47.42) | 43(53.75) | 62(51.24) | 50(55.56) | |
| No | 112(100) | 19(100) | 05(3.10) | 24(30.77) | 112(52.58) | 37(46.25) | 59(48.76) | 40(44.44) | |
| rs1458038 | FGF5 | ||||||||
| Yes | - | - | 156(97.50) | 55(70.51) | 101(47.42) | 45(56.25) | 62(51.24) | 49(54.44) | |
| No | 112(100) | 19(100) | 04(2.50) | 23(29.49) | 112(52.58) | 35(43.75) | 59(48.76) | 41(45.56) | |
| rs16960228 | PRKCA | ||||||||
| Yes | - | - | 158(98.75) | 56(71.79) | 102(47.89) | 47(58.75) | 63(52.07) | 48(53.33) | |
| No | 112(100) | 19(100) | 02(1.25) | 22(28.21) | 111(52.11) | 33(41.25) | 58(47.93) | 42(46.67) | |
| rs17010902 | APOA5 | ||||||||
| Yes | - | - | 152(95.00) | 54(69.23) | 98(46.01) | 44(55.00) | 61(50.41) | 47(52.22) | |
| No | 112(100) | 19(100) | 08(5.00) | 24(30.77) | 115(53.99) | 36(45.00) | 60(49.59) | 43(47.78) | |
| rs2106809 | ACE2 | ||||||||
| Yes | - | - | 157(98.10) | 54(69.23) | 103(48.36) | 45(56.25) | 62(51.24) | 50(55.56) | |
| No | 112(100) | 19(100) | 03(1.90) | 24(30.77) | 110(51.64) | 35(43.75) | 59(48.76) | 40(44.44) | |
| rs2107614 | WNK1 | ||||||||
| Yes | - | - | 155(96.90) | 55(70.51) | 100(46.95) | 47(58.75) | 62(51.24) | 46(51.11) | |
| No | 112(100) | 19(100) | 05(3.10) | 23(29.49) | 113(53.05) | 33(41.25) | 59(48.76) | 44(48.89) | |
| rs2269879 | DOT1L | ||||||||
| Yes | - | - | 156(97.50) | 54(69.23) | 102(47.89) | 44(55.00) | 63(52.07) | 49(54.44) | |
| No | 112(100) | 19(100) | 04(2.50) | 24(30.77) | 111(52.11) | 36(45.00) | 58(47.93) | 41(45.56) | |
| rs2277869 | WNK1 | ||||||||
| Yes | - | - | 156(97.50) | 55(70.51) | 101(47.42) | 46(57.50) | 61(50.41) | 49(54.44) | |
| No | 112(100) | 19(100) | 04(2.50) | 23(29.49) | 112(52.58) | 34(42.50) | 60(49.59) | 41(45.56) | |
| rs2400707 | ADRB2 | ||||||||
| Yes | - | - | 157(98.10) | 56(71.79) | 101(47.42) | 45(56.25) | 63(52.07) | 49(54.44) | |
| No | 112(100) | 19(100) | 03(1.90) | 22(28.21) | 112(52.58) | 35(43.75) | 58(47.93) | 41(45.56) | |
| rs2776546 | CSMD1 | ||||||||
| Yes | - | - | 158(98.75) | 56(71.79) | 102(47.89) | 47(58.75) | 63(52.07) | 48(53.33) | |
| No | 112(100) | 19(100) | 02(1.25) | 22(28.21) | 111(52.11) | 33(41.25) | 58(47.93) | 42(46.67) | |
| rs292449 | NEDD4L | ||||||||
| Yes | - | - | 156(97.50) | 55(70.51) | 101(47.42) | 45(56.25) | 63(2.07) | 48(53.33) | |
| No | 112(100) | 19(100) | 04(2.50) | 23(29.49) | 112(52.58) | 35(43.75) | 58(47.93) | 42(46.67) | |
| rs3184504 | SH2B3 | ||||||||
| Yes | - | - | 159(99.40) | 56(71.79) | 103(48.36) | 47(58.75) | 63(52.07) | 49(54.44) | |
| No | 112(100) | 19(100) | 01(0.60) | 22(28.21) | 110(51.64) | 33(41.25) | 58(47.93) | 41(45.56) | |
| rs4149601 | NEDD4L | ||||||||
| Yes | - | - | 159(99.40) | 56(71.79) | 103(48.36) | 47(58.75) | 63(52.07) | 49(54.44) | |
| No | 112(100) | 19(100) | 01(0.60) | 22(28.21) | 110(51.64) | 33(41.25) | 58(47.93) | 41(45.56) | |
| rs4551053 | EBF1 | ||||||||
| Yes | - | - | 160(100) | 56(71.79) | 104(48.83) | 47(58.75) | 63(52.07) | 50(55.56) | |
| No | 112(100) | 19(100) | - | 22(28.21) | 109(51.17) | 33(41.25) | 58(47.93) | 40(44.44) | |
| rs4791040 | PRKCA | ||||||||
| Yes | - | - | 110(68.75) | 38(48.72) | 72(33.80) | 30(37.50) | 42(34.71) | 38(42.22) | |
| No | 112(100) | 19(100) | 50(31.25) | 40(51.28) | 141(66.20) | 50(62.50) | 79(65.29) | 52(57.78) | |
| rs5051 | AGT | ||||||||
| Yes | - | - | 117(73.10) | 41(52.56) | 76(35.68) | 31(38.75) | 48(39.67) | 38(42.22) | |
| No | 112(100) | 19(100) | 43(26.90) | 37(47.44) | 137(64.32) | 49(61.25) | 73(60.33) | 52(57.78) | |
| rs6083538 | ZNF343 | ||||||||
| Yes | - | - | 156(97.50) | 56(71.79) | 100(46.95) | 46(57.50) | 63(52.07) | 47(52.22) | |
| No | 112(100) | 19(100) | 04(2.50) | 22(28.21) | 113(53.05) | 34(42.50) | 58(47.93) | 43(47.78) | |
| rs2070744 | NOS3 | ||||||||
| Yes | 112(100) | 19(100) | - | 22(28.21) | 109(51.17) | 33(41.25) | 58(47.93) | 40(44.44) | |
| No | - | - | 160(100) | 56(71.79) | 104(48.83) | 47(58.75) | 63(52.07) | 50(55.56) | |
| rs7297610 | YEATS4 | ||||||||
| Yes | 112(100) | 19(100) | - | 22(28.21) | 109(51.17) | 33(41.25) | 58(47.93) | 40(44.44) | |
| No | - | - | 160(100) | 56(71.79) | 104(48.83) | 47(58.75) | 63(52.07) | 50(55.56) | |
| dbSNP | Nucleotide Substitution | Feature | MAF (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Xhosa | Swati and Zulu | Yoruba | Luhya | Mexican | British | Punjabi | |||
| rs11189015 | C > G | Intron | 33.5 | - | 29.6 | 29.8 | 3.9 | 6.6 | 14.1 |
| rs1458038 | C > T | Intergenic | 20.3 | - | 94.4 | 97.5 | 73.4 | 73.6 | 77.1 |
| rs16960228 | C > T | Intron | 2.2 | - | 40.7 | 28.8 | 9.4 | 7.7 | 0.5 |
| rs17010902 | A > G | Intergenic | 59.5 | - | 0.5 | 3.5 | 26.0 | 8.8 | 16.1 |
| rs2106809 | A > G | Intron | 88.5 | - | 7.3 | 7.8 | 35.0 | 25.7 | 40.3 |
| rs2107614 | T > C | Intron | 53.5 | - | 38.9 | 53.0 | 64.8 | 73.1 | 69.3 |
| rs2269879 | C > T | Intron | 32.2 | - | 64.4 | 63.1 | 75.8 | 93.4 | 91.1 |
| rs2277869 | T > C | Non-coding exon | 20.5 | - | 13.0 | 17.7 | 18.0 | 14.3 | 19.3 |
| rs2400707 | A > G | 5 prime UTR | 37.26 | - | 56.9 | 43.3 | 82.8 | 60.4 | 70.8 |
| rs2776546 | C > A | Regulatory region | 48.7 | - | 41.2 | 40.4 | 26.6 | 17.0 | 13.0 |
| rs292449 | G > C | 5 prime UTR | 49.0 | - | 45.4 | 47.0 | 46.9 | 70.3 | 56.8 |
| rs3184504 | C > G | Missense | 100 | - | 100 | 100 | 22.7 | 50.0 | 7.3 |
| rs4149601 | G > A | Splice region | 51.5 | - | 37.5 | 50.0 | 11.7 | 31.3 | 18.8 |
| rs4551053 | G > A | Regulatory region | 13.6 | - | 11.1 | 11.6 | 16.4 | 33.0 | 43.2 |
| rs4791040 | T > C | Intron | 38.6 | - | 40.7 | 28.8 | 10.7 | 7.7 | 3.1 |
| rs5051 | C > T | Intron | 95.7 | - | 94.4 | 89.4 | 68.8 | 36.8 | 62.0 |
| rs6083538 | C > T | Intron | 15.0 | - | 7.9 | 9.1 | 46.9 | 47.8 | 37.5 |
| rs2070744 | C > T | Intron | - | 14.7 | 12.5 | 14.1 | 27.3 | 46.3 | 25.5 |
| rs7297610 | Intergenic | - | 52.4 | 37.0 | 31.3 | 5.5 | 6.0 | 2.1 | |
| dbSNP | Uncontrolled HPT (n; %) | Controlled HPT (n; %) | Unadjusted Odds Ratios (95% CI) | p-Value | Adjusted Odds Ratios (95% CI) | p-Value | Bonferroni-Adjusted p-Values |
|---|---|---|---|---|---|---|---|
| All | 31(19.37) | 129(80.63) | |||||
| rs11189015 | |||||||
| Genotypes | |||||||
| CC | 11(84.62) | 02(15.38) | 1 | 1 | |||
| GG | 52(76.47) | 16(23.53) | 1.01(0.25–4.03) | 0.982 | 0.82(0.30–2.22) | 0.699 | 0.041 |
| CG | 62(83.78) | 12(16.22) | 1.14(0.49–2.62) | 0.78(0.14–4.35) | 0.786 | 0.046 | |
| Alleles | |||||||
| G | 166(79.05) | 44(20.95) | 1 | 1 | |||
| C | 84(84.00) | 16(16.00) | 1.01(0.55–1.83) | 0.969 | 1.65(0.40–6.70) | 0.484 | 0.028 |
| rs1458038 | |||||||
| Genotypes | |||||||
| CC | 89(81.65) | 20(18.35) | 1 | 1 | |||
| TT | 06(66.67) | 03(33.33) | 0.85(0.34–2.10) | 0.727 | 2.32(0.32–16.87) | 0.403 | 0.023 |
| CT | 39(86.67) | 06(13.33) | 0.47(0.10–2.25) | 0.352 | 1.59(0.24–10.18) | 0.622 | 0.036 |
| Alleles | |||||||
| C | 217(82.51) | 46(17.49) | 1 | 1 | |||
| T | 51(80.95) | 12(19.05) | 1.02(0.50–2.08) | 0.939 | 0.85(0.19–3.74) | 0.832 | 0.048 |
| rs16960228 | |||||||
| Genotypes | |||||||
| CC | 123(81.46) | 28(18.54) | 1 | 1 | |||
| TT | - | - | - | - | |||
| TC | 04(57.14) | 03(42.86) | 2.19(0.51–9.32) | 0.286 | 0.29(0.46–1.91) | 0.201 | 0.011 |
| Alleles | |||||||
| T | 04(57.14) | 03(42.86) | 1 | 1 | |||
| C | 250(80.91) | 59(19.09) | 1.42(0.54–3.76) | 0.470 | 4.11(0.74–22.58) | 0.104 | 0.006 |
| rs17010902 | |||||||
| Genotypes | |||||||
| GG | 44(86.27) | 07(13.73) | 1 | 1 | |||
| AA | 17(77.27) | 05(22.73) | 1.59(0.63–3.99) | 0.45(0.98–2.12) | 0.318 | 0.018 | |
| AG | 61(77.22) | 18(22.78) | 1.33(0.44–4.01) | 0.46(0.06–3.67) | 0.471 | 0.027 | |
| Alleles | |||||||
| A | 95(77.24) | 28(22.76) | 1 | 1 | |||
| G | 149(82.32) | 32(17.68) | 1.37(0.77–2.42) | 0.275 | 0.34(0.57–2.10) | 0.249 | 0.014 |
| rs2106809 | |||||||
| Genotypes | |||||||
| GG | 09(75.00) | 03(25.00) | 1 | 1 | |||
| AA | 109(81.95) | 24(18.05) | 0.75(0.13–4.25) | 0.745 | 0.52(0.88–3.08) | 0.472 | 0.027 |
| AG | 09(75.00) | 03(25.00) | 1.36(0.34–5.32) | 0.657 | 0.40(0.75–2.16) | 0.290 | 0.017 |
| Alleles | |||||||
| G | 27(75.00) | 09(25.00) | 1 | 1 | |||
| A | 227(81.65) | 51(18.35) | 1.48(0.65–3.34) | 0.342 | 0.73(0.10–5.04) | 0.752 | 0.044 |
| rs2107614 | |||||||
| Genotypes | |||||||
| CC | 24(80.00) | 06(20.00) | 1 | 1 | |||
| TT | 33(80.49) | 08(19.51) | 1.06(0.38–3.00) | 0.901 | 1.17(0.37–3.68) | 0.786 | 0.046 |
| TC | 58(78.38) | 16(21.62) | 1.04(0.41–2.66) | 0.923 | 1.04(0.23–4.57) | 0.952 | 0.056 |
| Alleles | |||||||
| C | 106(79.10) | 28(20.90) | 1 | 1 | |||
| T | 124(79.49) | 32(20.51) | 1.01(0.57–1.77) | 0.970 | 6.69(1.42–31.55) | 0.016 | 0.0009 |
| rs2269879 | |||||||
| Genotypes | |||||||
| CC | 60(83.33) | 12(16.67) | 1 | 1 | |||
| TT | 12(75.00) | 04(25.00) | 1.48(0.64–3.45) | 0.357 | 0.63(0.11–3.44) | 0.599 | 0.035 |
| CT | 53(77.94) | 15(22.06) | 0.92(0.26–3.23) | 0.896 | 1.09(0.19–6.16) | 0.918 | 0.054 |
| Alleles | |||||||
| C | 173(81.60) | 39(18.40) | 1 | 1 | |||
| T | 77(77.00) | 23(23.00) | 0.75(0.42–1.34) | 0.342 | 1.49(0.33–6.65) | 0.597 | 0.035 |
| rs2277869 | |||||||
| Genotypes | |||||||
| CC | 03(75.00) | 01(25.00) | 1 | 1 | |||
| TT | 77(80.21) | 19(19.79) | 0.70(0.06–7.41) | 0.769 | 0.46(0.02–7.88) | 0.599 | 0.035 |
| CT | 45(80.36) | 11(19.64) | 0.97(0.42–2.22) | 0.948 | 1.06(0.38–2.92) | 0.899 | 0.052 |
| Alleles | |||||||
| C | 51(79.69) | 13(20.31) | 1 | 1 | |||
| T | 199(80.24) | 49(19.76) | 1.03(0.52–2.05) | 0.921 | 0.32(0.06–1.64) | 0.174 | 0.010 |
| rs2400707 | |||||||
| GG | 37(68.52) | 17(31.48) | 1 | 1 | |||
| AA | 22(88.00) | 03(12.00) | 0.36(0.15–0.85) | 0.020 | 0.84(0.16–4.28) | 0.842 | 0.049 |
| AG | 67(85.90) | 11(14.10) | 1.24(0.31–4.83) | 0.757 | 0.27(0.05–1.30) | 0.105 | 0.006 |
| Alleles | |||||||
| G | 141(75.81) | 45(24.19) | 1 | 1 | |||
| A | 111(86.72) | 17(13.28) | 7.34(3.05–17.67) | <0.0001 | 0.14(0.03–0.66) | 0.013 | 0.0007 |
| rs2776546 | |||||||
| Genotypes | |||||||
| AA | 32(82.05) | 07(17.95) | 1 | 1 | |||
| CC | 29(82.86) | 06(17.14) | 1.28(0.48–3.38) | 0.611 | 0.91(0.28–2.90) | 0.878 | 0.051 |
| CA | 66(78.57) | 18(21.43) | 1.36(0.49–3.78) | 0.551 | 1.12(0.28–4.40) | 0.862 | |
| Alleles | |||||||
| A | 130(80.25) | 32(19.75) | 1 | 1 | |||
| C | 124(80.52) | 30(19.48) | 1.01(0.58–1.77) | 0.951 | 3.78(1.04–13.74) | 0.043 | 0.0025 |
| rs292449 | |||||||
| Genotypes | |||||||
| GG | 29(74.36) | 10(25.64) | 1 | 1 | |||
| CC | 35(83.33) | 07(16.67) | 0.68(0.27–1.73) | 0.427 | 0.80(0.24–2.68) | 0.723 | 0.043 |
| GC | 61(81.33) | 14(18.67) | 1.24(0.46–3.36) | 0.665 | 0.59(0.16–2.41) | 0.422 | |
| Alleles | |||||||
| G | 119(77.78) | 34(22.22) | 1 | 1 | |||
| C | 131(82.39) | 28(17.61) | 1.33(0.76–2.33) | 0.308 | 0.34(0.09–1.21) | 0.097 | 0.005 |
| rs3184504 | |||||||
| Genotype | |||||||
| CC | 128(80.50) | 31(19.50) | - | - | - | - | |
| Alleles | |||||||
| C | 256(80.50) | 62(19.50) | - | - | - | - | |
| rs4149601 | |||||||
| Genotypes | |||||||
| AA | 34(85.00) | 06(15.00) | 1 | 1 | |||
| GG | 27(77.14) | 08(22.86) | 0.88(0.34–2.29) | 0.806 | 0.59(0.17–2.07) | 0.417 | 0.024 |
| GA | 67(79.76) | 17(20.24) | 1.43(0.51–3.98) | 0.485 | 0.41(0.96–1.74) | 0.228 | 0.013 |
| Alleles | |||||||
| A | 135(82.32) | 29(17.68) | 1 | 1 | |||
| G | 121(78.57) | 33(21.43) | 1.27(0.72–2.21) | 0.400 | 1.98(0.52–7.52) | 0.312 | 0.018 |
| rs4551053 | |||||||
| Genotypes | |||||||
| GG | 95(78.51) | 26(21.49) | 1 | 1 | |||
| AA | 01(100) | - | - | - | |||
| AG | 33(86.84) | 05(13.16) | 0.55(0.19–1.57) | 0.272 | 2.30(0.65–8.12) | 0.714 | 0.042 |
| Alleles | |||||||
| G | 223(79.64) | 57(20.36) | 1 | 1 | |||
| A | 35(87.50) | 05(12.50) | 1.78(0.67–4.77) | 0.245 | 0.17(0.01–2.00) | 0.162 | 0.009 |
| rs4791040 | |||||||
| Genotypes | |||||||
| TT | 44(86.27) | 07(13.73) | 1 | 1 | |||
| CC | 19(73.08) | 07(26.92) | 0.87(0.32–2.15) | 0.713 | 0.97(0.21–4.37) | 0.970 | 0.057 |
| TC | 26(78.79) | 07(21.21 | 1.42(0.53–3.75) | 0.478 | 0.50(0.08–2.84) | 0.437 | 0.025 |
| Alleles | |||||||
| T | 114(84.44) | 21(15.56) | 1 | 1 | |||
| C | 64(75.29) | 21(24.71) | 1.78(0.90–3.50) | 0.095 | 0.10(0.01–0.60) | 0.018 | 0.0007 |
| rs5051 | |||||||
| Genotypes | |||||||
| CC | - | - | - | - | |||
| TT | 88(82.24) | 19(17.76) | 1 | 1 | |||
| CT | 09(90.00) | 01(10.00) | 0.88(0.34–2.24) | 0.796 | 1.24(0.39–3.95) | 0.714 | 0.042 |
| Alleles | |||||||
| C | 9(90.00) | 01(10.00) | 1 | 1 | |||
| T | 185(82.59) | 39(17.41) | 0.52(0.06–4.28) | 0.549 | 1.90(0.15–24.22) | 0.618 | 0.036 |
| rs6083538 | |||||||
| Genotypes | |||||||
| CC | 94(82.46) | 20(17.54) | 1 | 1 | |||
| TT | 02(50.00) | 02(50.00) | 1.44(0.59–3.51) | 0.419 | 0.63(0.07–5.64) | 0.686 | 0.040 |
| CT | 29(76.32) | 09(23.68) | 1.08(0.91–6.19) | 0.926 | 0.99(0.13–7.56) | 0.994 | 0.058 |
| Alleles | |||||||
| C | 217(81.58) | 49(18.42) | 1 | 1 | |||
| T | 33(71.74) | 13(28.26) | 0.57(0.28–1.16) | 0.126 | 2.55(0.56–11.52) | 0.221 | 0.013 |
| dbSNP | Uncontrolled HPT (n; %) | Controlled HPT (n; %) | Unadjusted Odds Ratios (95% CI) | p-Value | Adjusted Odds Ratios (95% CI) | p-Value | Bonferroni-Adjusted p-Value |
|---|---|---|---|---|---|---|---|
| All | 71(54.19) | 60(45.80) | |||||
| rs2070744 | |||||||
| Genotypes | |||||||
| TT | 53(55.79) | 42(44.21) | 1 | 1 | |||
| CC | 02(40.00) | 03(60.00) | 4.22(1.15–15.47) | 0.030 | 10.44(2.16–50.29) | 0.003 | 0.0015 |
| TC | 16(51.61) | 15(48.39) | 0.10(0.02–0.48) | 0.004 | 38.76(5.54270.76) | 0.00023 | 0.0001 |
| Alleles | |||||||
| T | 122(55.71) | 99(44.29) | 1 | 1 | |||
| C | 20(45.55) | 21(57.45) | 0.77(0.39–1.50) | 0.449 | 1.68(0.82–3.42) | 0.151 | 0.076 |
| rs7297610 | |||||||
| Genotypes | |||||||
| CC | 27(54.00) | 23(46.00) | 1 | 1 | |||
| TT | 22(47.83) | 24(52.17) | 0.44(0.18–1.07) | 0.07 | 2.28(8.55–6.11) | 0.99 | 0.495 |
| CT | 21(61.76) | 13(38.24) | 0.40(0.16–0.98) | 0.045 | 0.94(0.37–2.34) | 0.898 | 0.449 |
| Alleles | |||||||
| C | 75(55.97) | 59(44.03) | 1 | 1 | |||
| T | 65(51.59) | 61(48.41) | 0.60(0.36–0.98) | 0.043 | 1.86(1.09–3.14) | 0.023 | 0.011 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masilela, C.; Pearce, B.; Ongole, J.J.; Adeniyi, O.V.; Benjeddou, M. Genomic Association of Single Nucleotide Polymorphisms with Blood Pressure Response to Hydrochlorothiazide among South African Adults with Hypertension. J. Pers. Med. 2020, 10, 267. https://doi.org/10.3390/jpm10040267
Masilela C, Pearce B, Ongole JJ, Adeniyi OV, Benjeddou M. Genomic Association of Single Nucleotide Polymorphisms with Blood Pressure Response to Hydrochlorothiazide among South African Adults with Hypertension. Journal of Personalized Medicine. 2020; 10(4):267. https://doi.org/10.3390/jpm10040267
Chicago/Turabian StyleMasilela, Charity, Brendon Pearce, Joven Jebio Ongole, Oladele Vincent Adeniyi, and Mongi Benjeddou. 2020. "Genomic Association of Single Nucleotide Polymorphisms with Blood Pressure Response to Hydrochlorothiazide among South African Adults with Hypertension" Journal of Personalized Medicine 10, no. 4: 267. https://doi.org/10.3390/jpm10040267