Metabolomics for Diagnosis and Prognosis of Uterine Diseases? A Systematic Review
Abstract
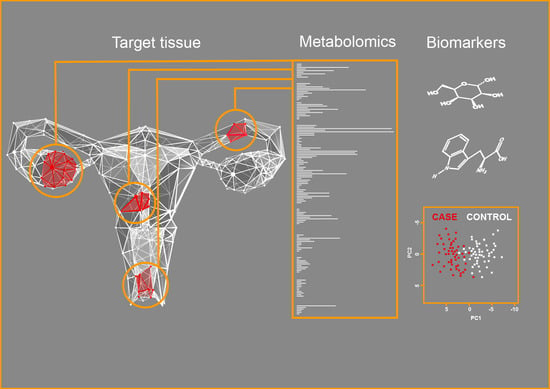
:1. Introduction
1.1. Rationale for the Review
1.2. Metabolomics
1.2.1. Nontargeted and Targeted Metabolomics
1.2.2. Mass Spectrometry
1.2.3. Nuclear Magnetic Resonance Spectroscopy
1.3. Uterine Diseases
1.3.1. Uterine Fibroids
1.3.2. Adenomyosis
1.3.3. Endometriosis
1.3.4. Cervical Cancer
1.3.5. Endometrial Cancer
2. Objectives
3. Methods
3.1. Information Sources, Search Strategy, and Eligibility Criteria
3.2. Data Extraction
3.3. Assessment of Risk of Bias
4. Results
4.1. Study Selection
4.2. Study Characteristics
4.2.1. Metabolomics in Uterine Fibroids and Endometriosis
| Sample | Method | Findings/Models | Reference |
| Uterine fibroids | |||
| tissue samples | LC-MS/MS | Leiomyomas/myometrium: 70 metabolites dysregulated; ↓ homocarnosine, haeme, biliverdin, stratification between FH and MED12 subtype | [67] |
| Endometriosis | |||
| serum | 1H-NMR | ↑ lactate, 2-hydroxybutyrate, succinate, Lys, glcerophosphocholine, citric acid, pyruvate, adipic acid; ↓ Ile, Leu, Arg, Asp, Ala, Glu, creatine; IW/E AUC: 0.99, SEN: 100%, SP: 91.6% | [69] |
| plasma | LC-MS/MS | ↑ lauroylcarnitine, oleylcarnitine, myristoylcarnitine, long-chain acylcarnitines; ↓ trimethylamine-N-oxide; CP/E: SEN: 81.8%, SP: 88.9%, PPV: 75% | [77] |
| peritoneal fluid | ESI-MS/MS | HW/OE (C0/PCae C36:0, PCaa C30:0/PCae C32:2, age) AUC: 0.94, SEN: 82.8%, SP: 94.4% | [72] |
| endometrial fluid | LC-MS/MS | ↓ sphingolipids, glycerolipids; ↑ mono or polyunsaturated TAG; CW/OE (model of 123 metabolites): SEN: 58.3%, SP: 100% | [73] |
| eutopic endometrium | LC-MS | IW/E (PC (18:1/22:6), PC (20:1/14:1), PC (20:3/20:4), PS (20:3/23:1), PA (25:5/22:6)); AUC: 0.87, SEN: 90.5%, SP: 75.0% | [75] |
| eutopic endometrium | LC-MS | IW/E (uric acid, hypoxanthine, lysophosphatidylethanolamine); AUC: 0.87, SEN: 66.7%, SP: 90.0% | [76] |
| Cervical cancer | |||
| plasma | LC-MS | CR/SD: (Val + Trp) AUC: 0.94, SEN: 87%, SP: 80%; CR + PR/SD: (Trp) AUC: 0.82 | [78] |
| plasma | LC-MS/MS | HW/CC: (bilirubin, LysoPC (17:0), n-oleoyl Thr, 12-hydroxydodecanoic acid, tetracosahexaenoic acid); AUC: 0.99, SEN: 98%, SP: 99% | [79] |
| plasma | LC-MS | phthalic acid, D-maltose, PG (12:0/13:0), LacCer (d18:1/16:0), PC (15:0/16:09); CC before/poor prognosis: AUC: 0.97, SEN: 94%, SP: 87%; CC before/good prognosis: AUC: 0.97, SEN: 92%, SP: 89% | [80] |
| Endometrial cancer | |||
| plasma | LC-MS/MS | BD/EC (C16/PCae C40:1, Pro/Tyr, PCaa C42:0/PCae C44:5); AUC: 0.84, SEN: 85.3%, SP: 69.2% MI-/MI+ (SMOH C14:1/SMOH C24:1, PCaa C40:2/PCaa C42:6); AUC: 0.86, SEN: 81.3%, SP: 86.4% LVI-/LVI+ (PCaa C34:4/PCae C38:3, C16:2/PCaa C38:1); AUC: 0.94, SEN: 88.9%, SP: 84.3% | [81] |
| plasma | LC-MS/MS | EC long/short survival (MetSO, serotonin, spermine, C3-OH, PCaa C36:5, SM C20:2, spermidine, C4:1, lyso PCaa C18:2, lysoPCaa C24:0, Asp, dimethylarginin, hexose, PCae C30:1); AUC: 0.965 | [82] |
| serum | GC-MS | EC type 1/type 2: ↓ lactic acid, cystine, Ser, malate, Glu, homocysteine; ↑ progesterone; Accuracy: 0.93, SEN: 96%, SP: 86% | [83] |
| serum | LC-MS/MS | C/EC (spermine, isovalerate, glycylvaline, gamma-glutamyl-2-aminobutyrate); AUC: 0.92 R/NR (2-oleoylgycerol, TAG 42:2-FA12:0); AUC: 0.90 | [84] |
| cervico-vaginal fluid | 1H-NMR | CP/EC (phosphocholine, malate, Asp); Accuracy: 0.78, SEN: 75%, SP: 80% | [85] |
4.2.2. Metabolomics in Cervical and Endometrial Cancers
4.3. Quality of the Selected Studies
5. Discussion
5.1. Main Findings
5.1.1. Contribution to Identification of Diagnostic and Prognostic Biomarkers
5.1.2. Contribution to Deciphering Pathophysiologies
5.2. Limitations of the Included Studies
5.3. Challenges and Future Perspectives
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [PubMed]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar]
- Vouk, K.; Hevir, N.; Ribic-Pucelj, M.; Haarpaintner, G.; Scherb, H.; Osredkar, J.; Moller, G.; Prehn, C.; Rizner, T.L.; Adamski, J. Discovery of phosphatidylcholines and sphingomyelins as biomarkers for ovarian endometriosis. Hum. Reprod. 2012, 27, 2955–2965. [Google Scholar]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [PubMed]
- Tweeddale, H.; Notley-McRobb, L.; Ferenci, T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (‘metabolome’) analysis. J. Bacteriol. 1998, 180, 5109–5116. [Google Scholar] [PubMed]
- Tokarz, J.; Haid, M.; Cecil, A.; Prehn, C.; Artati, A.; Möller, G.; Adamski, J. Endocrinology meets metabolomics: Achievements, pitfalls, and challenges. Trends Endocrinol. Metab. 2017, 28, 705–721. [Google Scholar] [PubMed] [Green Version]
- Hocher, B.; Adamski, J. Metabolomics for clinical use and research in chronic kidney disease. Nat. Rev. Nephrol. 2017, 13, 269–284. [Google Scholar]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar]
- Koal, T.; Deigner, H.-P. Challenges in Mass Spectrometry Based Targeted Metabolomics. Current Mol. Med. 2010, 10, 216–226. [Google Scholar]
- Naz, S.; Vallejo, M.; García, A.; Barbas, C. Method validation strategies involved in non-targeted metabolomics. J. Chrom. A 2014, 1353, 99–105. [Google Scholar]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Hoffmann, E. Tandem mass spectrometry: A primer. J. Mass Spectromet. 1996, 31, 129–137. [Google Scholar] [CrossRef]
- Fuhrer, T.; Heer, D.; Begemann, B.; Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011, 83, 7074–7080. [Google Scholar] [CrossRef] [PubMed]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrometr. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef]
- Haid, M.; Muschet, C.; Wahl, S.; Römisch-Margl, W.; Prehn, C.; Möller, G.; Adamski, J. Long-term stability of human plasma metabolites during storage at −80 °C. J. Proteome Res. 2018, 17, 203–211. [Google Scholar] [CrossRef]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Lubbe, A.; Ali, K.; Verpoorte, R.; Choi, Y.H. NMR-based metabolomics analysis. In Metabolomics in Practice; Lämmerhofer, M., Weckwerth, W., Eds.; Wiley-VCH: Weinheim, Germany, 2013. [Google Scholar]
- Schripsema, J. Application of NMR in plant metabolomics: Techniques, problems and prospects. Phytochem. Anal. 2010, 21, 14–21. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra- and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Malz, F.; Jancke, H. Validation of quantitative NMR. J. Pharm. Biomed Analys. 2005, 38, 813–823. [Google Scholar] [CrossRef]
- Baird, D.D.; Dunson, D.B.; Hill, M.C.; Cousins, D.; Schectman, J.M. High cumulative incidence of uterine leiomyoma in black and white women: Ultrasound evidence. Am. J. Obstet. Gynecol. 2003, 188, 100–107. [Google Scholar] [CrossRef]
- Naftalin, J.; Hoo, W.; Pateman, K.; Mavrelos, D.; Holland, T.; Jurkovic, D. How common is adenomyosis? A prospective study of prevalence using transvaginal ultrasound in a gynaecology clinic. Hum. Reprod. 2012, 27, 3432–3439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay , J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Vlahos, N.F.; Theodoridis, T.D.; Partsinevelos, G.A. Myomas and Adenomyosis: Impact on Reproductive Outcome. Biomed. Res. Int. 2017, 2017, 5926470. [Google Scholar] [CrossRef] [PubMed]
- Donnez, J.; Dolmans, M.M. Uterine fibroid management: From the present to the future. Hum. Reprod. Update 2016, 22, 665–686. [Google Scholar] [CrossRef]
- Miller, D.S.; King, L.P. Gynecologic oncology group trials in uterine corpus malignancies: Recent progress. J. Gynecol. Oncol. 2008, 19, 218–222. [Google Scholar] [CrossRef] [Green Version]
- El-Balat, A.; DeWilde, R.L.; Schmeil, I.; Tahmasbi-Rad, M.; Bogdanyova, S.; Fathi, A.; Becker, S. Modern Myoma Treatment in the Last 20 Years: A Review of the Literature. Biomed. Res. Int. 2018, 2018, 4593875. [Google Scholar] [CrossRef]
- Torres-de la Roche, L.A.; Becker, S.; Cezar, C.; Hermann, A.; Larbig, A.; Leicher, L.; Di Spiezio Sardo, A.; Tanos, V.; Wallwiener, M.; Verhoeven, H.; et al. Pathobiology of myomatosis uteri: The underlying knowledge to support our clinical practice. Arch. Gynecol. Obstet. 2017, 296, 701–707. [Google Scholar] [CrossRef]
- Vannuccini, S.; Petraglia, F. Recent advances in understanding and managing adenomyosis. F1000Research 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Chapron, C.; Vannuccini, S.; Santulli, P.; Abrão, M.S.; Carmona, F.; Fraser, I.S.; Gordts, S.; Guo, S.W.; Just, P.A.; Noël, J.C.; et al. Diagnosing adenomyosis: An integrated clinical and imaging approach. Hum. Reprod. Update. 2020, 26, 392–411. [Google Scholar] [CrossRef]
- Yeniel, O.; Cirpan, T.; Ulukus, M.; Ozbal, A.; Gundem, G.; Ozsener, S.; Zekioglu, O.; Yilmaz, H. Adenomyosis: Prevalence, risk factors, symptoms and clinical findings. Clin. Exp. Obstet. Gynecol. 2007, 34, 163–167. [Google Scholar] [PubMed]
- Leyendecker, G.; Bilgicyildirim, A.; Inacker, M.; Stalf, T.; Huppert, P.; Mall, G.; Böttcher, B.; Wildt, L. Adenomyosis and endometriosis. Re-visiting their association and further insights into the mechanisms of auto-traumatisation. An MRI study. Arch. Gynecol. Obstet. 2015, 291, 917–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannuccini, S.; Tosti, C.; Carmona, F.; Huang, S.J.; Chapron, C.; Guo, S.W.; Petraglia, F. Pathogenesis of adenomyosis: An update on molecular mechanisms. Reprod. Biomed. Online 2017, 35, 592–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, J.; Sterrenburg, M.; Lane, S.; Maheshwari, A.; Li, T.C.; Cheong, Y. Reproductive, obstetric, and perinatal outcomes of women with adenomyosis and endometriosis: A systematic review and meta-analysis. Hum. Reprod. Update 2019, 25, 592–632. [Google Scholar] [CrossRef] [PubMed]
- Vannuccini, S.; Luisi, S.; Tosti, C.; Sorbi, F.; Petraglia, F. Role of medical therapy in the management of uterine adenomyosis. Fertil. Steril. 2018, 109, 398–405. [Google Scholar] [CrossRef] [Green Version]
- Giudice, L.C.; Kao, L.C. Endometriosis. Lancet 2004, 364, 1789–1799. [Google Scholar] [CrossRef]
- Bulun, S.E.; Yilmaz, B.D.; Sison, C.; Miyazaki, K.; Bernardi, L.; Liu, S.; Kohlmeier, A.; Yin, P.; Milad, M.; Wei, J. Endometriosis. Endocr. Rev. 2019, 40, 1048–1079. [Google Scholar] [CrossRef]
- Vercellini, P.; Viganò, P.; Somigliana, E.; Fedele, L. Endometriosis: Pathogenesis and treatment. Nat. Rev. Endocrinol. 2014, 10, 261–275. [Google Scholar] [CrossRef]
- Burney, R.O.; Giudice, L.C. Pathogenesis and pathophysiology of endometriosis. Fertil. Steril. 2012, 98, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Rogers, P.A.; Adamson, G.D.; Al-Jefout, M.; Becker, C.M.; D’Hooghe, T.M.; Dunselman, G.A.; Fazleabas, A.; Giudice, L.C.; Horne, A.W.; Hull, M.L.; et al. Research Priorities for Endometriosis. Reprod. Sci. 2017, 24, 202–226. [Google Scholar] [CrossRef] [Green Version]
- Simoens, S.; Dunselman, G.; Dirksen, C.; Hummelshoj, L.; Bokor, A.; Brandes, I.; Brodszky, V.; Canis, M.; Colombo, G.L.; DeLeire, T.; et al. The burden of endometriosis: Costs and quality of life of women with endometriosis and treated in referral centres. Human Reprod. (Oxf. Engl.) 2012, 27, 1292–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nnoaham, K.E.; Hummelshoj, L.; Webster, P.; d’Hooghe, T.; de Cicco Nardone, F.; de Cicco Nardone, C.; Jenkinson, C.; Kennedy, S.H.; Zondervan, K.T.; World Endometriosis Research Foundation Global Study of Women’s Health consortium. Impact of endometriosis on quality of life and work productivity: A multicenter study across ten countries. Fertil. Steril. 2011, 96, 366–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisolle, M.; Donnez, J. Peritoneal endometriosis, ovarian endometriosis, and adenomyotic nodules of the rectovaginal septum are three different entities. Fertil. Steril. 1997, 68, 585–596. [Google Scholar] [CrossRef]
- Andrews, W.C.; Buttram, V.C.J.; Weed, J.; Hammond, C.; Thomas, H. Revised American Fertility Society classification of endometriosis: 1985. Fertil. Steril. 1985, 43, 351–352. [Google Scholar]
- Johnson, N.P.; Hummelshoj, L.; Consortium, W.E.S.M. Consensus on current management of endometriosis. Hum. Reprod. (Oxf. Engl.) 2013, 28, 1552–1568. [Google Scholar] [CrossRef] [PubMed]
- Nap, A.W. Theories on the Pathogenesis of Endometriosis. In Endometriosis Science and Practice; Giudice, L.C., Evers, J.L., Healy, D.L., Eds.; Wiley-Blackwell A John Wiley and Sons, Ltd. Publication: Singapore, 2012; pp. 42–53. [Google Scholar]
- Cousins, F.L.; O, D.F.; Gargett, C.E. Endometrial stem/progenitor cells and their role in the pathogenesis of endometriosis. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 50, 27–38. [Google Scholar] [CrossRef]
- Chan, C.K.; Aimagambetova, G.; Ukybassova, T.; Kongrtay, K.; Azizan, A. Human Papillomavirus Infection and Cervical Cancer: Epidemiology, Screening, and Vaccination-Review of Current Perspectives. J. Oncol. 2019, 2019, 3257939. [Google Scholar] [CrossRef]
- National Cancer Institute (US). Cervical Cancer Treatment (PDQ). NCBI Bookshelf, A Service of the National Library of Medicine; National Institutes of Health: Bethesda, MD, USA, 2019. [Google Scholar]
- Faridi, R.; Zahra, A.; Khan, K.; Idrees, M. Oncogenic potential of Human Papillomavirus (HPV) and its relation with cervical cancer. Virol. J. 2011, 8, 269. [Google Scholar] [CrossRef] [Green Version]
- Kessler, T.A. Cervical Cancer: Prevention and Early Detection. Semin. Oncol. Nurs. 2017, 33, 172–183. [Google Scholar] [CrossRef]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Lee, Y.C.; Lheureux, S.; Oza, A.M. Treatment strategies for endometrial cancer: Current practice and perspective. Curr. Opin. Obstet. Gynecol. 2017, 29, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.M.; Orr, J.; Leitao, M.; Salom, E.; Gehrig, P.; Olawaiye, A.B.; Brewer, M.; Boruta, D.; Villella, J.; Herzog, T.; et al. Endometrial cancer: A review and current management strategies: Part I. Gynecol. Oncol. 2014, 134, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.; Soslow, R.A. Updates in diagnostic immunohistochemistry in endometrial carcinoma. Semin. Diagn. Pathol. 2014, 31, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.L.; Mutter, G.L. Molecular and pathologic aspects of endometrial carcinogenesis. J. Clin. Oncol. 2006, 24, 4783–4791. [Google Scholar] [CrossRef]
- Ryan, A.J.; Susil, B.; Jobling, T.W.; Oehler, M.K. Endometrial cancer. Cell Tissue Res. 2005, 322, 53–61. [Google Scholar] [CrossRef]
- Amant, F.; Moerman, P.; Neven, P.; Timmerman, D.; Van Limbergen, E.; Vergote, I. Endometrial cancer. Lancet 2005, 366, 491–505. [Google Scholar] [CrossRef]
- Meyer, L.A.; Broaddus, R.R.; Lu, K.H. Endometrial cancer and Lynch syndrome: Clinical and pathologic considerations. Cancer Control. 2009, 16, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Bracewell-Milnes, T.; Saso, S.; Abdalla, H.; Nikolau, D.; Norman-Taylor, J.; Johnson, M.; Holmes, E.; Thum, M.Y. Metabolomics as a tool to identify biomarkers to predict and improve outcomes in reproductive medicine: A systematic review. Hum. Reprod. Update 2017, 23, 723–736. [Google Scholar] [CrossRef] [Green Version]
- Siristatidis, C.S.; Sertedaki, E.; Vaidakis, D.; Varounis, C.; Trivella, M. Metabolomics for improving pregnancy outcomes in women undergoing assisted reproductive technologies. Cochrane Database Syst. Rev. 2018, 3, CD011872. [Google Scholar] [CrossRef]
- McInnes, M.D.F.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Lumbreras, B.; Porta, M.; Márquez, S.; Pollán, M.; Parker, L.A.; Hernández-Aguado, I. QUADOMICS: An adaptation of the Quality Assessment of Diagnostic Accuracy Assessment (QUADAS) for the evaluation of the methodological quality of studies on the diagnostic accuracy of ‘-omics’-based technologies. Clin Biochem. 2008, 41, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, H.-R.; Mehine, M.; Mäkinen, N.; Pasanen, A.; Pitkänen, E.; Karhu, A.; Sarvilinna, N.S.; Sjöberg, J.; Heikinheimo, O.; Bützow, R.; et al. Global metabolomic profiling of uterine leiomyomas. Br. J. Cancer 2017, 117, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Dutta, M.; Joshi, M.; Srivastava, S.; Lodh, I.; Chakravarty, B.; Chaudhury, K. A metabonomics approach as a means for identification of potential biomarkers for early diagnosis of endometriosis. Mol. Biosyst. 2012, 8, 3281–3287. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.K.; Dutta, M.; Joshi, M.; Srivastava, S.; Chakravarty, B.; Chaudhury, K. 1H NMR based targeted metabolite profiling for understanding the complex relationship connecting oxidative stress with endometriosis. Biomed. Res. Int. 2013, 2013, 329058. [Google Scholar] [CrossRef] [PubMed]
- Braga, D.P.A.F.; Montani, D.A.; Setti, A.S.; Turco, E.G.L.; Oliveira-Silva, D.; Borges, E. Metabolomic profile as a noninvasive adjunct tool for the diagnosis of Grades III and IV endometriosis-related infertility. Mol. Reprod. Dev. 2019, 86, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Munoz, S.; Morcillo, I.; Puchades-Carrasco, L.; Paya, V.; Pellicer, A.; Pineda-Lucena, A. Nuclear magnetic resonance metabolomic profiling of urine provides a noninvasive alternative to the identification of biomarkers associated with endometriosis. Fertil. Steril. 2015, 104, 1202–1209. [Google Scholar] [CrossRef] [Green Version]
- Vouk, K.; Ribič-Pucelj, M.; Adamski, J.; Rižner, T.L. Altered levels of acylcarnitines, phosphatidylcholines, and sphingomyelins in peritoneal fluid from ovarian endometriosis patients. J. Steroid Biochem. Mol. Biol. 2016, 159, 60–69. [Google Scholar] [CrossRef]
- Domínguez, F.; Ferrando, M.; Díaz-Gimeno, P.; Quintana, F.; Fernández, G.; Castells, I.; Simón, C. Lipidomic profiling of endometrial fluid in women with ovarian endometriosis. Biol. Reprod. 2017, 96, 772–779. [Google Scholar] [CrossRef]
- Chagovets, V.V.; Wang, Z.; Kononikhin, A.S.; Starodubtseva, N.L.; Borisova, A.; Salimova, D.; Popov, I.A.; Kozachenko, A.V.; Chingin, K.; Chen, H.; et al. Endometriosis foci differentiation by rapid lipid profiling using tissue spray ionization and high resolution mass spectrometry. Sci. Rep. 2017, 7, 2546. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gao, Y.; Guan, L.; Zhang, H.; Sun, J.; Gong, X.; Li, D.; Chen, P.; Ma, Z.; Liang, X.; et al. Discovery of Phosphatidic Acid, Phosphatidylcholine, and Phosphatidylserine as Biomarkers for Early Diagnosis of Endometriosis. Front. Physiol. 2018, 9, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Guan, L.; Zhang, H.; Gao, Y.; Sun, J.; Gong, X.; Li, D.; Chen, P.; Liang, X.; Huang, M.; et al. Endometrium metabolomic profiling reveals potential biomarkers for diagnosis of endometriosis at minimal-mild stages. Reprod. Biol. Endocrinol. 2018, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Letsiou, S.; Peterse, D.P.; Fassbender, A.; Hendriks, M.M.; van den Broek, N.J.; Berger, R.; Dorien, O.F.; Vanhie, A.; Vodolazkaia, A.; Van Langendonckt, A.; et al. Endometriosis is associated with aberrant metabolite profiles in plasma. Fertil. Steril. 2017, 107, 699–706.e696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Yin, M.; Sun, F.; Zhang, T.; Zhou, X.; Li, H.; Zheng, J.; Chen, X.; Li, C.; Ning, X.; et al. A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol. Biosyst. 2014, 10, 2126–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Xia, B.; Wang, W.; Cheng, J.; Yin, M.; Xie, H.; Li, J.; Ma, L.; Yang, C.; Li, A.; et al. A Comprehensive Analysis of Metabolomics and Transcriptomics in Cervical Cancer. Sci. Rep. 2017, 7, 43353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, Q.; Wang, T.; Liang, H.; Wang, Y.; Duan, Y.; Song, M.; Wang, Y.; Jin, H. Prognostic biomarkers of cervical squamous cell carcinoma identified via plasma metabolomics. Medicine (Baltimore) 2019, 98, e16192. [Google Scholar] [CrossRef]
- Knific, T.; Vouk, K.; Smrkolj, Š.; Prehn, C.; Adamski, J.; Rižner, T.L. Models including plasma levels of sphingomyelins and phosphatidylcholines as diagnostic and prognostic biomarkers of endometrial cancer. J. Steroid Biochem. Mol. Biol. 2018, 178, 312–321. [Google Scholar] [CrossRef]
- Strand, E.; Tangen, I.L.; Fasmer, K.E.; Jacob, H.; Halle, M.K.; Hoivik, E.A.; Delvoux, B.; Trovik, J.; Haldorsen, I.S.; Romano, A.; et al. Blood Metabolites Associate with Prognosis in Endometrial Cancer. Metabolites 2019, 9, 302. [Google Scholar] [CrossRef] [Green Version]
- Troisi, J.; Sarno, L.; Landolfi, A.; Scala, G.; Martinelli, P.; Venturella, R.; Di Cello, A.; Zullo, F.; Guida, M. Metabolomic Signature of Endometrial Cancer. J. Proteome Res. 2018, 17, 804–812. [Google Scholar] [CrossRef]
- Audet-Delage, Y.; Villeneuve, L.; Grégoire, J.; Plante, M.; Guillemette, C. Identification of Metabolomic Biomarkers for Endometrial Cancer and Its Recurrence after Surgery in Postmenopausal Women. Front. Endocrinol. (Lausanne) 2018, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.C.; Chen, K.; Chiu, C.Y.; Lu, K.Y.; Lu, H.Y.; Chiang, M.H.; Tsai, C.K.; Lo, C.J.; Cheng, M.L.; Chang, T.C.; et al. Metabolomic biomarkers in cervicovaginal fluid for detecting endometrial cancer through nuclear magnetic resonance spectroscopy. Metabolomics 2019, 15, 146. [Google Scholar] [CrossRef] [PubMed]
- Abudula, A.; Rouzi, N.; Xu, L.; Yang, Y.; Hasimu, A. Tissue-based metabolomics reveals potential biomarkers for cervical carcinoma and HPV infection. Bosn. J. Basic Med. Sci. 2020, 20, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.S.; Jung, S.; Han, J.S.; Hwang, G.S.; Kim, M.K. LC/MS-Based Polar Metabolite Profiling Identified Unique Biomarker Signatures for Cervical Cancer and Cervical Intraepithelial Neoplasia Using Global and Targeted Metabolomics. Cancers 2019, 11, 511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilhan, Z.E.; Łaniewski, P.; Thomas, N.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Deciphering the complex interplay between microbiota, H.P.V; inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine 2019, 44, 675–690. [Google Scholar] [CrossRef] [Green Version]
- Tokareva, A.O.; Chagovets, V.V.; Starodubtseva, N.L.; Nazarova, N.M.; Nekrasova, M.E.; Kononikhin, A.S.; Frankevich, V.E.; Nikolaev, E.N.; Sukhikh, G.T. Feature selection for OPLS discriminant analysis of cancer tissue lipidomics data. J. Mass Spectrom. 2020, 55, e4457. [Google Scholar] [CrossRef]
- Yin, M.Z.; Tan, S.; Li, X.; Hou, Y.; Cao, G.; Li, K.; Kou, J.; Lou, G. Identification of phosphatidylcholine and lysophosphatidylcholine as novel biomarkers for cervical cancers in a prospective cohort study. Tumour Biol. 2016, 37, 5485–5492. [Google Scholar] [CrossRef]
- Ye, N.; Liu, C.; Shi, P. Metabolomics analysis of cervical cancer, cervical intraepithelial neoplasia and chronic cervicitis by 1H NMR spectroscopy. Eur. J. Gynaecol. Oncol. 2015, 36, 174–180. [Google Scholar]
- Schuler, K.M.; Rambally, B.S.; DiFurio, M.J.; Sampey, B.P.; Gehrig, P.A.; Makowski, L.; Bae-Jump, V.L. Antiproliferative and metabolic effects of metformin in a preoperative window clinical trial for endometrial cancer. Cancer Med. 2015, 4, 161–173. [Google Scholar] [CrossRef]
- Altadill, T.; Dowdy, T.M.; Gill, K.; Reques, A.; Menon, S.S.; Moiola, C.P.; Lopez-Gil, C.; Coll, E.; Matias-Guiu, X.; Cabrera, S.; et al. Metabolomic and Lipidomic Profiling Identifies The Role of the RNA Editing Pathway in Endometrial Carcinogenesis. Sci. Rep. 2017, 7, 8803. [Google Scholar] [CrossRef]
- Shi, K.; Wang, Q.; Su, Y.; Xuan, X.; Liu, Y.; Chen, W.; Qian, Y.; Lash, G.E. Identification and functional analyses of differentially expressed metabolites in early stage endometrial carcinoma. Cancer Sci. 2018, 109, 1032–1043. [Google Scholar] [CrossRef] [Green Version]
- Audet-Delage, Y.; Grégoire, J.; Caron, P.; Turcotte, V.; Plante, M.; Ayotte, P.; Simonyan, D.; Villeneuve, L.; Guillemette, C. Estradiol metabolites as biomarkers of endometrial cancer prognosis after surgery. J. Steroid Biochem. Mol. Biol. 2018, 178, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahado-Singh, R.O.; Lugade, A.; Field, J.; Al-Wahab, Z.; Han, B.; Mandal, R.; Bjorndahl, T.C.; Turkoglu, O.; Graham, S.F.; Wishart, D.; et al. Metabolomic prediction of endometrial cancer. Metabolomics 2017, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Ihata, Y.; Miyagi, E.; Numazaki, R.; Muramatsu, T.; Imaizumi, A.; Yamamoto, H.; Yamakado, M.; Okamoto, N.; Hirahara, F. Amino acid profile index for early detection of endometrial cancer: Verification as a novel diagnostic marker. Int. J. Clin. Oncol. 2014, 19, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Cummings, M.; Massey, K.A.; Mappa, G.; Wilkinson, N.; Hutson, R.; Munot, S.; Saidi, S.; Nugent, D.; Broadhead, T.; Wright, A.I.; et al. Integrated eicosanoid lipidomics and gene expression reveal decreased prostaglandin catabolism and increased 5-lipoxygenase expression in aggressive subtypes of endometrial cancer. J. Pathol. 2019, 247, 21–34. [Google Scholar] [CrossRef]
- Jové, M.; Gatius, S.; Yeramian, A.; Portero-Otin, M.; Eritja, N.; Santacana, M.; Colas, E.; Ruiz, M.; Pamplona, R.; Matias-Guiu, X. Metabotyping human endometrioid endometrial adenocarcinoma reveals an implication of endocannabinoid metabolism. Oncotarget 2016, 7, 52364–52374. [Google Scholar] [CrossRef] [Green Version]
- Eritja, N.; Jové, M.; Fasmer, K.E.; Gatius, S.; Portero-Otin, M.; Trovik, J.; Krakstad, C.; Sol, J.; Pamplona, R.; Haldorsen, I.S.; et al. Tumour-microenvironmental blood flow determines a metabolomic signature identifying lysophospholipids and resolvin D as biomarkers in endometrial cancer patients. Oncotarget 2017, 8, 109018–109026. [Google Scholar] [CrossRef]
- Gaudet, M.M.; Falk, R.T.; Stevens, R.D.; Gunter, M.J.; Bain, J.R.; Pfeiffer, R.M.; Potischman, N.; Lissowska, J.; Peplonska, B.; Brinton, L.A.; et al. Analysis of serum metabolic profiles in women with endometrial cancer and controls in a population-based case-control study. J. Clin. Endocrinol. Metab. 2012, 97, 3216–3223. [Google Scholar] [CrossRef]
- Lee, Y.H.; Tan, C.W.; Venkatratnam, A.; Tan, C.S.; Cui, L.; Loh, S.F.; Griffith, L.; Tannenbaum, S.R.; Chan, J.K. Dysregulated sphingolipid metabolism in endometriosis. J. Clin. Endocrinol. Metab. 2014, 99, E1913–E1921. [Google Scholar] [CrossRef] [Green Version]
- Starodubtseva, N.; Chagovets, V.; Borisova, A.; Salimova, D.; Aleksandrova, N.; Chingin, K.; Chen, H.; Frankevich, V. Identification of potential endometriosis biomarkers in peritoneal fluid and blood plasma via shotgun lipidomics. Clin. Mass Spectrom. 2019, 13, 21–26. [Google Scholar] [CrossRef]
- Santulli, P.; Marcellin, L.; Noël, J.C.; Borghese, B.; Fayt, I.; Vaiman, D.; Chapron, C.; Méhats, C. Sphingosine pathway deregulation in endometriotic tissues. Fertil. Steril. 2012, 97, 904–911. [Google Scholar] [CrossRef]
- Corre, I.; Niaudet, C.; Paris, F. Plasma membrane signaling induced by ionizing radiation. Mutat. Res. Rev. Mutat. Res. 2010, 704, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Hasim, A.; Ali, M.; Mamtimin, B.; Ma, J.Q.; Li, Q.Z.; Abudula, A. Metabonomic signature analysis of cervical carcinoma and precancerous lesions in women by (1)H NMR spectroscopy. Exp. Ther. Med. 2012, 3, 945–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasim, A.; Aili, A.; Maimaiti, A.; Mamtimin, B.; Abudula, A.; Upur, H. Plasma-free amino acid profiling of cervical cancer and cervical intraepithelial neoplasia patients and its application for early detection. Mol. Biol. Rep. 2013, 40, 5853–5859. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.; Wilson, D.J. A study of metabolites as intermediate effectors in angiogenesis. Angiogenesis 2001, 4, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Xie, G.; Jia, W. Metabonomics of human colorectal cancer: New approaches for early diagnosis and biomarker discovery. J. Proteome Res. 2014, 13, 3857–3870. [Google Scholar] [CrossRef] [PubMed]
- Njoku, K.; Chiasserini, D.; Whetton, A.D.; Crosbie, E.J. Proteomic biomarkers for the detection of endometrial cancer. Cancers 2019, 11, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, X.; Liang, D.; Zhao, A.; Jia, W.; Chen, T. MCEE 2.0: More options and enhanced performance. Anal. Bioanal. Chem. 2019, 411, 5089–5098. [Google Scholar] [CrossRef]
- Rizner, T.L.; Adamski, J. Paramount importance of sample quality in pre-clinical and clinical research-Need for standard operating procedures (SOPs). J. Steroid Biochem. Mol. Biol. 2019, 186, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Dunn, W.B.; Wilson, I.D.; Nicholls, A.W.; Broadhurst, D. The importance of experimental design and QC samples in large-scale and MS-driven untargeted metabolomic studies of humans. Bioanalysis 2012, 4, 2249–2264. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hoene, M.; Wang, X.; Yin, P.; Häring, H.U.; Xu, G.; Lehmann, R. Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Anal. Chim. Acta 2018, 1037, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Kastenmüller, G.; He, Y.; Belcredi, P.; Möller, G.; Prehn, C.; Mendes, J.; Wahl, S.; Roemisch-Margl, W.; Ceglarek, U.; et al. Differences between human plasma and serum metabolite profiles. PLoS ONE 2011, 6, e21230. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.M.; Kim, K.M.; Choi, M.H.; Jung, B.H.; Lee, J.; Kong, G.; Nam, S.J.; Kim, S.; Bai, S.W.; Chung, B.C. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta 2009, 400, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]





| Search Query | Search Results | Selected Manuscripts | Additional Manuscripts | Included |
|---|---|---|---|---|
| (myoma uteri) OR (uterine myoma) OR (uterine myomas) OR leiomyoma OR leiomyomas OR (uterine fibroid) OR (uterine fibroids) OR (uterus myomatosus) AND metabolomic AND (English[Language]) NOT review | 8 (Pubmed) 3 (OVID) 12 (Scopus) | 1 (Pubmed) 1 * (OVID) 0 (Scopus) | 0 | 1 |
| adenomyosis OR (endometriosis interna) AND metabolomic AND (English[Language]) NOT review | 1 (Pubmed) 0 (OVID) 1 (Scopus) | 0 (Pubmed) 0 (OVID) 0 (Scopus) | 0 | 0 |
| endometriosis AND (metabolomic OR metabolomics) AND (English[Language]) NOT review | 31 (Pubmed) 21 (OVID) 38 (Scopus) | 13 (Pubmed) 10 * (OVID) 4 */1 (Scopus) | 3 | 17 |
| (cervical cancer) OR (cervix cancer) OR (cervical carcinoma) AND (metabolomic OR metabolomics) AND English[Language]) NOT review | 81 (Pubmed) 21 (OVID) 96 (Scopus) | 9 (Pubmed) 5 * (OVID) 1 * (Scopus) | 3 | 12 |
| (endometrial cancer) OR (uterine cancer) OR (endometrial carcinoma) OR (uterine carcinoma) AND metabolomic AND (English[Language]) NOT review | 69 (Pubmed) 10 (OVID) 92 (Scopus) | 12 (Pubmed) 8 * (OVID) 2 * (Scopus) | 2 | 14 |
| Total | 190 (Pubmed) 55 (OVID) 239 (Scopus) | 35 (Pubmed) 24 * (OVID) 1/7 * (Scopus) | 8 | 44 |
| Inclusion criteria | research papers, papers in English, studies in humans, tissue or physiological fluids, except follicular fluid * |
| Exclusion criteria | review papers, papers in other languages, studies in animals, follicular fluid, studies including only unidentified metabolites, studies focusing on fertility *, studies in cell culture ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokarz, J.; Adamski, J.; Rižner, T.L. Metabolomics for Diagnosis and Prognosis of Uterine Diseases? A Systematic Review. J. Pers. Med. 2020, 10, 294. https://doi.org/10.3390/jpm10040294
Tokarz J, Adamski J, Rižner TL. Metabolomics for Diagnosis and Prognosis of Uterine Diseases? A Systematic Review. Journal of Personalized Medicine. 2020; 10(4):294. https://doi.org/10.3390/jpm10040294
Chicago/Turabian StyleTokarz, Janina, Jerzy Adamski, and Tea Lanišnik Rižner. 2020. "Metabolomics for Diagnosis and Prognosis of Uterine Diseases? A Systematic Review" Journal of Personalized Medicine 10, no. 4: 294. https://doi.org/10.3390/jpm10040294