“Cancer 2015”: A Prospective, Population-Based Cancer Cohort—Phase 1: Feasibility of Genomics-Guided Precision Medicine in the Clinic
Abstract
:1. Introduction
- Establishment of a database consisting of biospecimens as well as clinical and epidemiological data to be used as a clinical and research resource.
- To screen cancer tumour DNA isolated from diagnostic tissue samples from patients independent of cancer subtype, to determine if any mutations are present that may potentially be of clinical and therapeutic significance.
- To yield data on the total population frequency of these mutations and identify patients that may benefit from therapeutics targeted against these “actionable” mutations.
- To collect health-related quality of life (HRQoL) responses longitudinally, and link in administrative health care resource use and cost data, in order to facilitate the assessment of the value of targeted therapies, and cancer care more generally.
2. Results
2.1. Cohort Characteristics

| Characteristic | N | % |
|---|---|---|
| Total Consented: | 1685 | |
| Deceased | 233 | 13.8 |
| Withdrawn | 63 | 3.7 |
| Male | 916 | 54.0 |
| Female | 769 | 45.5 |
| Recruited from Institution: | ||
| Cabrini Hospital | 322 | 19.0 |
| Geelong Hospital | 284 | 16.7 |
| Peter MacCallum Cancer Centre | 523 | 30.9 |
| Royal Melbourne Hospital | 362 | 21.5 |
| Warrnambool Hospital | 194 | 11.5 |
| Regional Statistics: | ||
| Metropolitan | 936 | 57.7 (69) |
| Non-Metropolitan | 685 | 42.3 (31) |
| Age (years and 10-yr deciles) | ||
| Median | 63.2 | |
| 11–20 | 1 | 0.1 |
| 21–30 | 27 | 1.6 |
| 31–40 | 73 | 4.4 |
| 41–50 | 204 | 12.2 |
| 51–60 | 374 | 22.3 |
| 61–70 | 573 | 34.2 |
| 71–80 | 317 | 18.9 |
| 81–90 | 103 | 6.1 |
| 91–100 | 3 | 0.2 |
| Region of Origin (Birth): | ||
| Africa | 22 | 1.3 (2.5) |
| Asia | 70 | 4.2 (4.8) |
| Australia (inc. Oceania) | 1222 | 72.5 (67.6) |
| Europe | 268 | 15.9 (24.3) |
| North America | 9 | 0.5 (0.4) |
| South America | 2 | 0.1 (0.4) |
| Marital Status*: | ||
| Never married | 69 | 4.1 |
| Married | 803 | 47.7 |
| Divorced | 97 | 5.8 |
| Widowed | 108 | 6.4 |
| Separated | 19 | 1.1 |
| Not stated | 575 | 34.1 |
| Education Level*: | ||
| Primary | 51 | 3.0 |
| Junior Secondary | 168 | 10.0 |
| Senior Secondary | 148 | 8.8 |
| Graduate | 105 | 6.2 |
| Post-graduate | 44 | 2.6 |
| No formal education | 9 | 0.5 |
| Not stated | 1146 | 68.0 |
| Disease Presentation Mode: | ||
| Symptomatic | 1081 | 64.2 |
| Asymptomatic/incidental | 196 | 11.6 |
| Screening | 361 | 21.4 |
| Not Stated | 38 | 2.3 |
| Performance Status (ECOG): | ||
| 0 | 1066 | 63.3 |
| 1 | 400 | 23.7 |
| 2 | 121 | 7.2 |
| 3 | 42 | 2.5 |
| 4 | 4 | 0.2 |
| Charlson Co-Morbidities Index: | ||
| 0–5 | 1448 | 85.9 |
| 6–10 | 58 | 3.4 |
| >10 | 170 | 10.1 |
| Private Hospital Insurance: | ||
| Yes | 694 | 41.2 |
| No | 948 | 56.3 |
| Smoking Status: | ||
| Daily | 209 | 12.4 |
| Weekly | 13 | 0.8 |
| Irregular | 25 | 1.5 |
| Ex-smoker | 747 | 44.3 |
| Never Smoked | 630 | 37.4 |
| Past History of Cancer: | ||
| Yes | 303 | 18.0 |
| No | 1336 | 79.3 |
| Family History of Cancer (1st/2nd order blood relative): | ||
| Yes | 1106 | 65.6 |
| No | 507 | 30.1 |
| Hereditary Syndromes | 10 | 0.6 |
| Blood Samples Obtained: | ||
| Received | 1505 | 89.3 |
| Unavailable/Insufficient tissue | 172 | 10.2 |
| Quality of Life Questionnaire responses: | ||
| Baseline | 1606 | 95.3 |
| Follow-up | 1271 | 75.4 |
| Medicare/Pharmaceutical Benefit Scheme Co-Consent | 1590 | 94.4 |
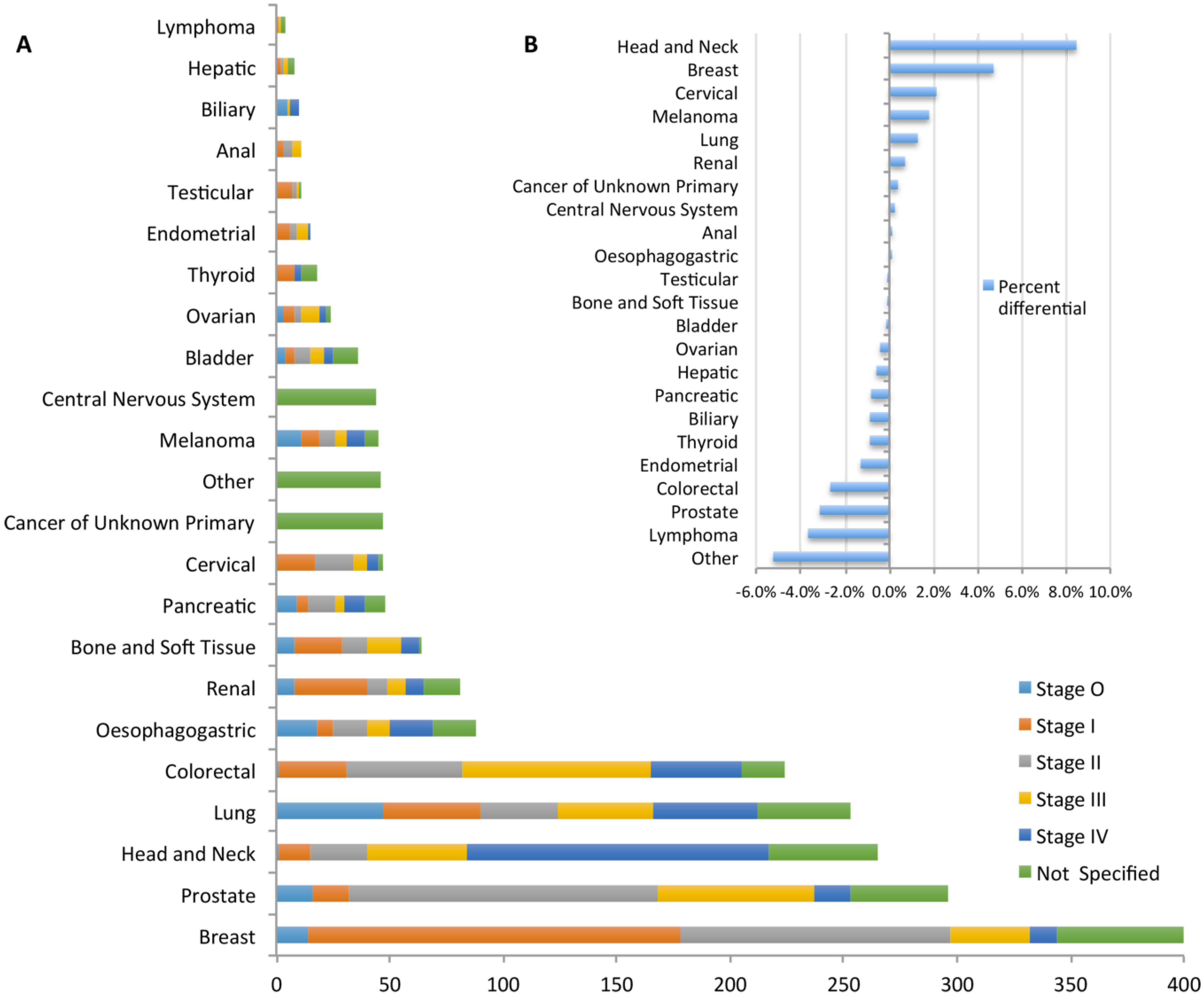
2.2. Data Completion
2.3. Genomic Assay of Biospecimens

2.4. Patient Follow Up

3. Discussion
4. Experimental Section
4.1. Study Design
4.2. Biospecimens
4.3. Data Collection
4.4. Data Linkages
4.5. Patient Follow up
5. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Stewart, B.W.; Wild, C. World Cancer Report 2014. International Agency for Research on Cancer; World Health Organization: Lyon, France, 2014. [Google Scholar]
- Cancer Council of Victoria Cancer Registry. Cancer in Victoria: Statistics and Trends. Available online: http://www.cancervic.org.au/research/registry-statistics/cancer-in-victoria (accessed on 21 September 2015).
- Callaway, E. Norway to bring cancer-gene tests to the clinic. Nature 2012. [Google Scholar] [CrossRef]
- Rabesandratana, R. U.K.’s 100,000 Genomes Project Gets £ 300 Million to Finish the Job by 2017. Available online: http://news.sciencemag.org/biology/2014/08/u-k-s-100000-genomes-project-gets-300-million-finish-job-2017 (accessed on 22 October 2015).
- The Academy of Medical Sciences. Realising the Potential of Stratified Medicine. July 2013. Available online: http://www.acmedsci.ac.uk/policy/policy-projects/Stratified-medicine/ (accessed on 22 October 2015).
- Australian Institute of Health and Welfare and Australasian Association of Cancer Registries 2012. Cancer in Australia: An Overview 2012; AIHW: Canberra, Australia, 2012. [Google Scholar]
- Health Data Standards Committee. Data Set Specification, National Health Data Dictionary version 12 Supplement. Available online: http://www.aihw.gov.au/publication-detail/?id=6442467641 (accessed on 21 September 2015).
- Oken, M.M.; Creech, R.H.; Tormey, D.C.; Horton, J.; Davis, T.E.; McFadden, E.T.; Carbone, P.P. Toxicity and response criteria of the eastern cooperative oncology group. Am. J. Clin. Oncol. 1982, 5, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Charlson, M.E.; Pompei, P.; Ales, K.L.; MacKenzie, C.R. A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J. Chronic Dis. 1987, 40, 373–383. [Google Scholar] [CrossRef]
- Wong, S.Q.; Li, J.; Tan, A.Y.-C.; Vedururu, R.; Pang, J.-M.B.; Do, H.; Ellul, J.; Doig, K.; Bell, A.; McArthur, G.A.; et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med. Genomics 2014. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Q.; Fellowes, A.; Doig, K.; Ellul, J.; Bosma, T.; Vedururu, R.; Tan, A.Y.-C.; Weiss, J.; Chan, K.S.; Lucas, M.; et al. Assessing the clinical value of targeted massively parallel sequencing in a longitudinal, prospective population-based study of cancer patients. Br. J. Cancer 2015, 112, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Chavez, A.; Thomas, A.; Rajan, A.; Raffeld, M.; Morrow, B.; Kelly, R.; Carter, C.A.; Guha, U.; Killian, K.; Lau, C.C.; et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: A biomarker-derived, multiarm, multihistology phase II basket trial. J. Clin. Oncol. 2015, 33, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Brusco, L.; Shaw, K.; Horombe, C.; Kopetz, S.; Davies, M.A.; Routbort, M.; Piha-Paul, S.A.; Janku, F.; Ueno, N.; et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J. Clin. Oncol. 2015, 33, 2753–2762. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Puzanove, I.; Subbiah, V.; Faris, J.E.; Chau, I.; Blay, J.-Y.; Wolf, J.; Raje, N.S.; Diamond, E.L.; Hollebecque, A.; et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 2015, 373, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Eng, K.; Mosquera, J.M.; Sigaras, A.; Romanel, A.; Rennart, H.; Kossai, M.; Pauli, C.; Faltas, B.; Fontugne, J.; et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015, 4, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Schott, A.F.; Perou, C.M.; Hayes, D.F. Genome medicine in cancer: What’s in a name? Cancer Res. 2015, 75, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, J. All aboard: Will molecular tumor boards help cancer patients? Nat. Med. 2015, 21, 655–656. [Google Scholar] [CrossRef] [PubMed]
- National Health and Medical Research Council. National Statement on Ethical Conduct in Human Research (2007). Available online: https://www.nhmrc.gov.au/guidelines-publications/e72 (accessed on 21 September 2015).
- Aaronson, N.K.; Ahmedzai, S.; Bergman, B.; Bullinger, M.; Cull, A.; Duez, N.J.; Filiberti, A.; Flechtner, H.; Fleishman, S.B.; de Haes, J.C.J.M.; et al. The European organization for research and treatment of cancer QLQ-C30: A quality-of-life instrument for use in international clinical trials in oncology. J. Natl. Cancer Inst. 1993, 85, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R. EuroQol: The current state of play. Health Policy 1996, 37, 53–72. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parisot, J.P.; Thorne, H.; Fellowes, A.; Doig, K.; Lucas, M.; McNeil, J.J.; Doble, B.; Dobrovic, A.; John, T.; James, P.A.; et al. “Cancer 2015”: A Prospective, Population-Based Cancer Cohort—Phase 1: Feasibility of Genomics-Guided Precision Medicine in the Clinic. J. Pers. Med. 2015, 5, 354-369. https://doi.org/10.3390/jpm5040354
Parisot JP, Thorne H, Fellowes A, Doig K, Lucas M, McNeil JJ, Doble B, Dobrovic A, John T, James PA, et al. “Cancer 2015”: A Prospective, Population-Based Cancer Cohort—Phase 1: Feasibility of Genomics-Guided Precision Medicine in the Clinic. Journal of Personalized Medicine. 2015; 5(4):354-369. https://doi.org/10.3390/jpm5040354
Chicago/Turabian StyleParisot, John P., Heather Thorne, Andrew Fellowes, Ken Doig, Mark Lucas, John J. McNeil, Brett Doble, Alexander Dobrovic, Thomas John, Paul A. James, and et al. 2015. "“Cancer 2015”: A Prospective, Population-Based Cancer Cohort—Phase 1: Feasibility of Genomics-Guided Precision Medicine in the Clinic" Journal of Personalized Medicine 5, no. 4: 354-369. https://doi.org/10.3390/jpm5040354
APA StyleParisot, J. P., Thorne, H., Fellowes, A., Doig, K., Lucas, M., McNeil, J. J., Doble, B., Dobrovic, A., John, T., James, P. A., Lipton, L., Ashley, D., Hayes, T., McMurrick, P., Richardson, G., Lorgelly, P., Fox, S. B., & Thomas, D. M. (2015). “Cancer 2015”: A Prospective, Population-Based Cancer Cohort—Phase 1: Feasibility of Genomics-Guided Precision Medicine in the Clinic. Journal of Personalized Medicine, 5(4), 354-369. https://doi.org/10.3390/jpm5040354






