A Gustatory Receptor GR8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Tissue Collection
2.2. Sugar Compounds
2.3. RNA Isolation and cDNA Synthesis
2.4. Gene Cloning and RACE Amplification
2.5. Sequence Analysis and Phylogenetic Tree Construction
2.6. Spatial and Temporal Expression Analysis
2.7. Vector Construction and cRNA Synthesis
2.8. Receptor Expression in Xenopus Oocytes and Electrophysiological Recordings
2.9. Proboscis Extension Reflex (PER) Assay
3. Results
3.1. Gene Cloning and Phylogenetic Analysis
3.2. Spatial and Temporal Expression of SlitGR8
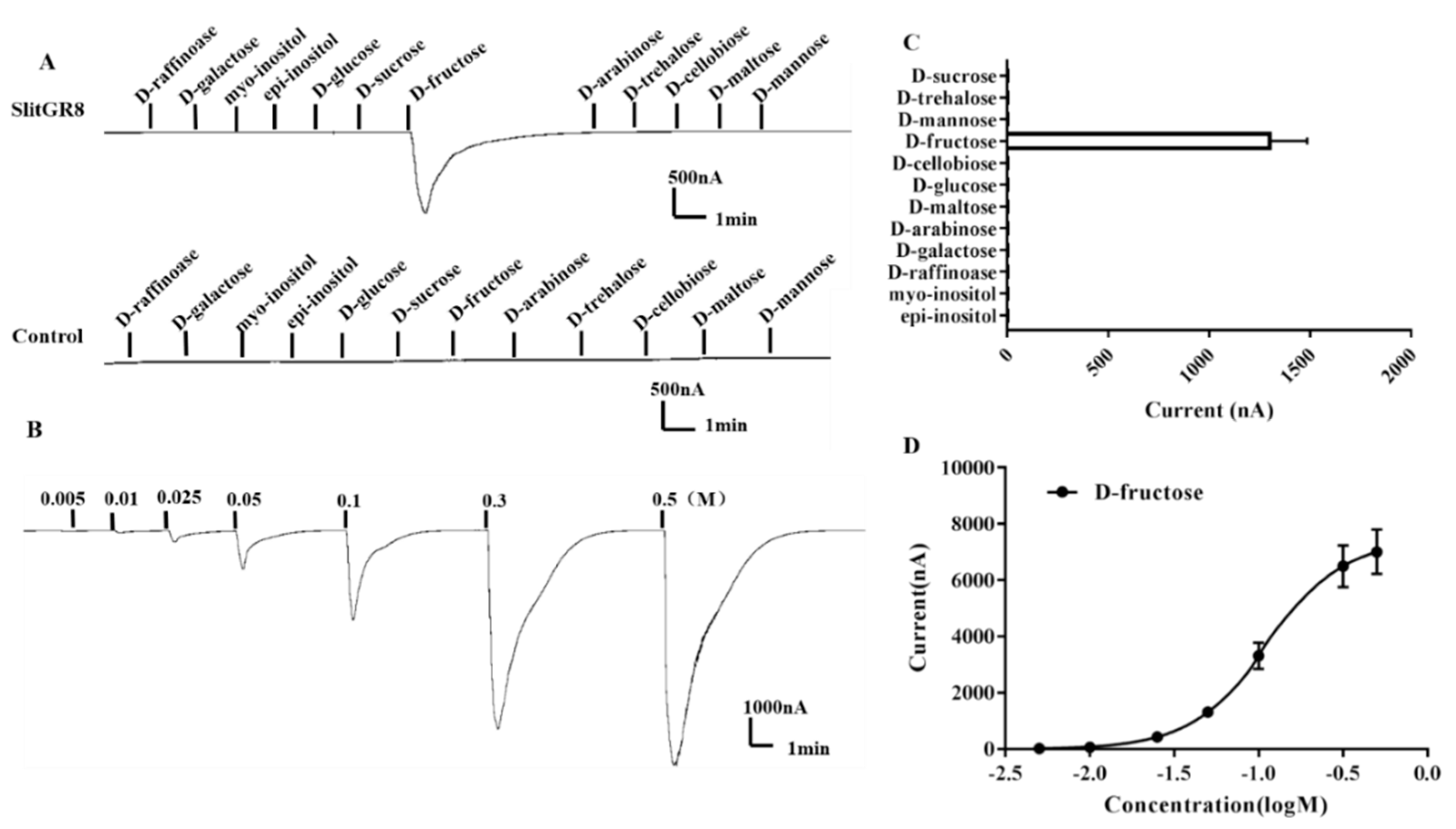
3.3. Functional Characterization of SlitGR8
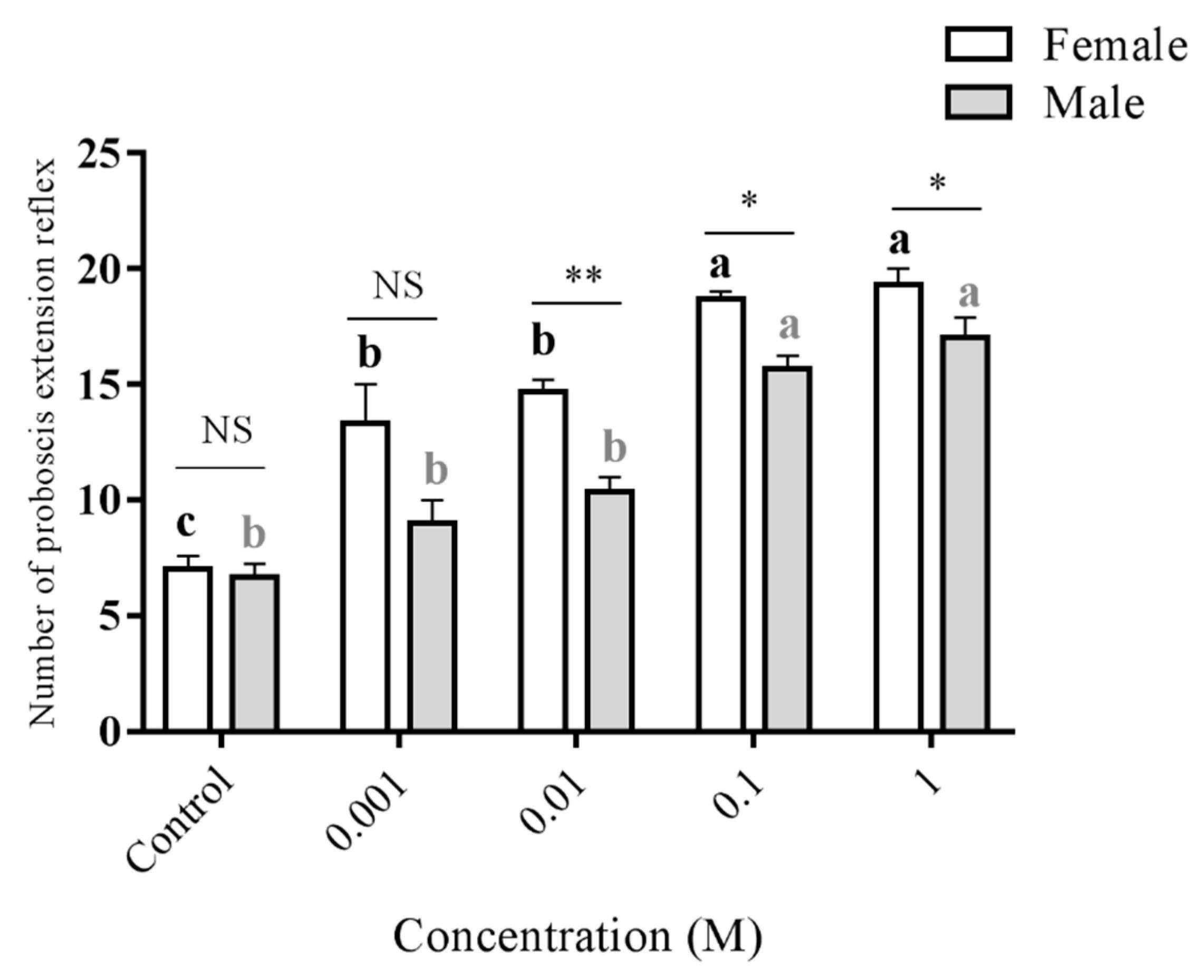
3.4. Proboscis Extension Reflex (PER) Induced by D-Fructose
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hanson, F.E.; Dethier, V.G. Rôle of gustation and olfaction in food plant discrimination in the tobacco hornworm, Manduca sexta. J. Insect Physiol. 1973, 19, 1019–1031. [Google Scholar] [CrossRef]
- Dethier, V.G. The importance of stimulus patterns for host-plant recognition and acceptance. In The Host-Plant in Relation to Insect Behaviour and Reproduction; Jermy, T., Ed.; Springer: Boston, MA, USA, 1976; pp. 67–70. ISBN 978-1-4613-4274-8. [Google Scholar]
- Stocker, R.F. The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 1994, 275, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Dahanukar, A.; Hallem, E.A.; Carlson, J.R. Insect chemoreception. Curr. Opin. Neurobiol. 2005, 15, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Sachse, S.; Michnick, S.W.; Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Anderson, A.R.; Trowell, S.C.; Luo, A.-R.; Xiang, Z.-H.; Xia, Q.-Y. Topological and functional characterization of an insect gustatory receptor. PLoS ONE 2011, 6, e24111. [Google Scholar]
- Schoonhoven, L.M.; Jermy, T.; van Loon, J.J.A. Insect-Plant Biology; Chapman & Hall: London, UK, 1998; pp. 183–189. [Google Scholar]
- Sato, K.; Tanaka, K.; Touhara, K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 11680–11685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, T.; Slone, J.; Song, X.; Amrein, H. A fructose receptor functions as a nutrient sensor in the Drosophila Brain. Cell 2012, 151, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.Y.; Dahanukar, A.; Weiss, L.A.; Carlson, J.R. The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. USA 2007, 104, 3574–3578. [Google Scholar] [CrossRef] [PubMed]
- Erdelyan, C.N.G.; Mahood, T.H.; Bader, T.S.Y.; Whyard, S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 2012, 21, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Moon, S.J.; Wang, X.; Ren, Q.; Montell, C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 2008, 18, 1797–1801. [Google Scholar] [CrossRef] [PubMed]
- Larsson, M.C.; Domingos, A.I.; Jones, W.D.; Chiappe, M.E.; Amrein, H.; Vosshall, L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 2004, 43, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Chyb, S.; Dahanukar, A.; Wickens, A.; Carlson, J.R. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl. Acad. Sci. USA 2003, 100, 14526–14530. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Moon, S.J.; Montell, C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA 2007, 104, 14110–14115. [Google Scholar] [CrossRef] [PubMed]
- Kikuta, S.; Endo, H.; Tomita, N.; Takada, T.; Morita, C.; Asaoka, K.; Sato, R. Characterization of a ligand-gated cation channel based on an inositol receptor in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 74, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.J.; Ning, C.; Guo, H.; Jia, Y.Y.; Huang, L.Q.; Qu, M.J.; Wang, C.Z. A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2015, 60, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, H.J.; Anderson, A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 2012, 38, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Huang, Y.; Du, J. Sex pheromones and reproductive behavior of Spodoptera litura (Fabricius) moths reared from larvae treated with four insecticides. J. Chem. Ecol. 2004, 30, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhu, L.; Ni, J.; Chao, X. A method of rearing the beet armyworm Spodoptera exigua. Kunchong Zhishi 2002, 39, 229–231. [Google Scholar]
- Cheng, T.; Wu, J.; Wu, Y.; Chilukuri, R.V.; Huang, L.; Yamamoto, K.; Feng, L.; Li, W.; Chen, Z.; Guo, H.; et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 2017, 1, 1747. [Google Scholar] [CrossRef] [PubMed]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Robertson, H.M. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 2008, 17, 621–629. [Google Scholar] [CrossRef]
- Liu, N.Y.; Xu, W.; Papanicolaou, A.; Dong, S.L.; Anderson, A. Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics 2014, 15, 597. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, G.; Carey, A.F.; Carlson, J.R.; Zwiebel, L.J. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2010, 107, 4418–4423. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Sun, S.J.; Khuhro, S.A.; Elzaki, M.E.A.; Yan, Q.; Dong, S.L. Functional characterization of pheromone receptors in the moth Athetis dissimilis (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 2019, 158, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Wykes, G.R. An investigation of the sugars present in the nectar of flowers of various species. New Phytol. 1952, 51, 210–215. [Google Scholar] [CrossRef]
- Pate, J.S.; Peoples, M.B.; Storer, P.J.; Atkins, C.A. The extrafloral nectaries of cowpea (Vigna unguiculata (L.) Walp.) II. Nectar composition, origin of nectar solutes, and nectary functioning. Planta 1985, 166, 28–38. [Google Scholar] [CrossRef]
- Heil, M. Nectar: Generation, regulation and ecological functions. Trends Plant Sci. 2011, 16, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.P.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef]
- Nishino, H.; Nishikawa, M.; Yokohari, F.; Mizunami, M. Dual, multilayered somatosensory maps formed by antennal tactile and contact chemosensory afferents in an insect brain. J. Comp. Neurol. 2005, 493, 291–308. [Google Scholar] [CrossRef] [PubMed]
- Popescu, A.; Couton, L.; Almaas, T.J.; Rospars, J.P.; Wright, G.A.; Marion-Poll, F.; Anton, S. Function and central projections of gustatory receptor neurons on the antenna of the noctuid moth Spodoptera littoralis. J. Comp. Physiol. A 2013, 199, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, P.G. Sugars, Amino Acids, and Ascorbic Acid as Phagostimulants for Larvae of Antitrogus parvulus and Lepidiota negatoria (Coleoptera: Scarabaeidae). J. Econ. Entomol. 1992, 85, 106–111. [Google Scholar] [CrossRef]
- Brouwers, E.V.M. Glucose/fructose ratio in the food of honeybee larvae during caste differentiation. J. Apic. Res. 1984, 23, 94–101. [Google Scholar] [CrossRef]
- Qin, J.D.; Li, L.Y.; Wei, D.Y.; Wang, Z.D. Some characteristics in the food preference and nutrition of the cotton bollworm. Acta Entomologica Sinica 1962, 11, 327–340. [Google Scholar]



| SlitGR8 | HarmGR4 | HarmGR9 | BmorGR9 | DmelGR43a | |
|---|---|---|---|---|---|
| NCBI-SlitGR8 | – | 93.3% | 94.5% | 60.1% | 23.9% |
| Paper-SlitGR8 | – | 91.9% | 95.1% | 61.1% | 24.4% |
| SlitGR8 | – | 87.3% | 89.8% | 57.6% | 23.3% |
| HarmGR4 | – | 95.0% | 59.2% | 23.7% | |
| HarmGR9 | – | 62.2% | 24.5% | ||
| BmorGR9 | – | 24.0% | |||
| DmelGR43a | – |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.-L.; Yan, Q.; Yang, Y.-L.; Hou, W.; Miao, C.-L.; Peng, Y.-C.; Dong, S.-L. A Gustatory Receptor GR8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura. Insects 2019, 10, 272. https://doi.org/10.3390/insects10090272
Liu X-L, Yan Q, Yang Y-L, Hou W, Miao C-L, Peng Y-C, Dong S-L. A Gustatory Receptor GR8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura. Insects. 2019; 10(9):272. https://doi.org/10.3390/insects10090272
Chicago/Turabian StyleLiu, Xiao-Long, Qi Yan, Yi-Lin Yang, Wen Hou, Chun-Li Miao, Ying-Chuan Peng, and Shuang-Lin Dong. 2019. "A Gustatory Receptor GR8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura" Insects 10, no. 9: 272. https://doi.org/10.3390/insects10090272





