The Genetic Diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the Phylogenetic Relationship of Scutigeromorpha Using the Mitochondrial Genome
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection, Species Identification and DNA Extraction
2.2. COX1 Sequences and Next Generation Sequencing
2.3. Sequence Analyses and Annotation
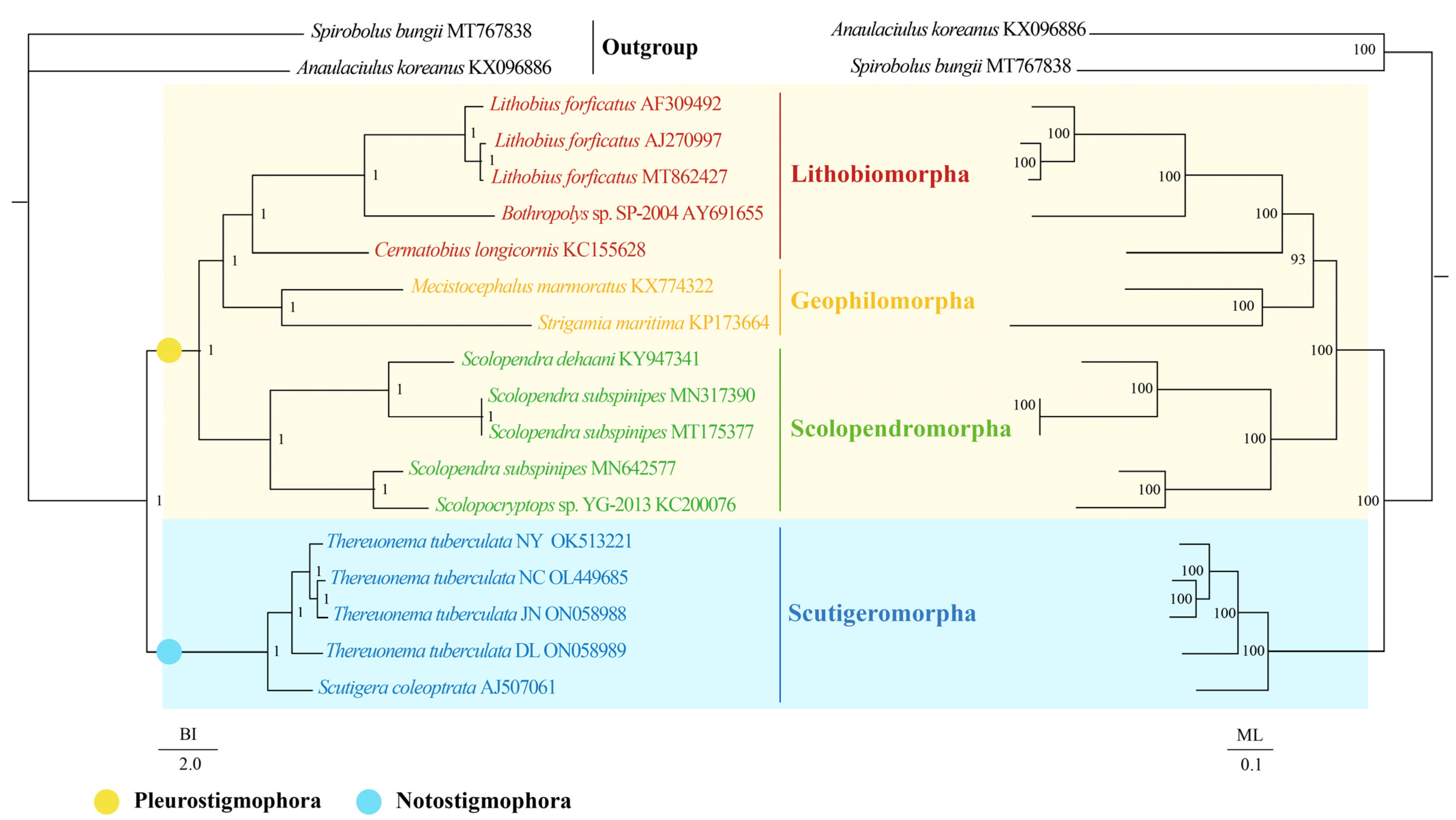
2.4. Phylogenetic Analyses
3. Results and Discussion
3.1. Species Identification
3.2. Mitochondrial Genome Organization and Composition
3.2.1. General Features of Mitochondrial Genomes
3.2.2. Protein-Coding Genes and Codon Usages
3.2.3. Ribosomal RNAs, Transfer RNAs and Hairpin Structures
3.2.4. A + T Rich Region
3.3. The Corrected Pairwise Genetic Distance of T. tuberculata
3.4. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edgecombe, G.D.; Giribet, G. Evolutionary biology of centipedes (Myriapoda: Chilopoda). Annu. Rev. Entomol. 2007, 52, 151–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgecombe, G.D. Centipede systematics: Progress and problems. Zootaxa 2007, 1668, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Bortolin, F.; Fusco, G.; Bonato, L. Comparative analysis of diet in syntopic geophilomorph species (Chilopoda, Geophilomorpha) using a DNA-based approach. Soil Biol. Biochem. 2018, 127, 223–229. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Giribet, G. Myriapod Phylogeny and the Relationships of Chilopoda. In Biodiversidad, Taxonomía y Biogeografia de Artrópodos de México: Hacia una Síntesis de su Conocimiento; Prensas de Ciencias, Universidad Nacional Autónoma de México: Mexico City, Mexico, 2002; Volume 3, pp. 143–168. [Google Scholar]
- Edgecombe, G.D.; Giribet, G. Adding mitochondrial sequence data (16S rRNA and cytochrome c oxidase subunit I) to the phylogeny of centipedes (Myriapoda: Chilopoda): An analysis of morphology and four molecular loci. J. Zool. Syst. Evol. Res. 2010, 42, 89–134. [Google Scholar] [CrossRef]
- Giribet, G.; Carranza, S.; Riutort, M.; Baguna, J.; Ribera, C. Internal phylogeny of the Chilopoda (Myriapoda, Arthropoda) using complete 18S rDNA and partial 28S rDNA sequences. Philos. Trans. R. Soc. Lond. B Biol. 1999, 354, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Murienne, J.; Edgecombe, G.D.; Giribet, G. Including secondary structure, fossils and molecular dating in the centipede tree of life. Mol. Phylogenet. Evol. 2010, 57, 301–313. [Google Scholar] [CrossRef]
- Giribet, G.; Edgecombe, G.D. Stable phylogenetic patterns in scutigeromorph centipedes (Myriapoda: Chilopoda: Scutigeromorpha): Dating the diversification of an ancient lineage of terrestrial arthropods. Invertebr. Syst. 2013, 27, 485–501. [Google Scholar] [CrossRef]
- Shear, W.; Jeram, A.; Selden, P. Centiped legs (Arthropoda, Chilopoda, Scutigeromorpha) from the Silurian and Devonian of Britain and the Devonian of North America. Am. Mus. Novit. 1998, 3231, 1–16. [Google Scholar]
- Wilson, H.M. First Mesozoic scutigeromorph centipede, from the lower Cretaceous of Brazil. Palaeontology 2001, 44, 489–495. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Giribet, G. A century later—A total evidence re-evaluation of the phylogeny of scutigeromorph centipedes (Myriapoda:Chilopoda). Invertebr. Syst. 2006, 20, 503–525. [Google Scholar] [CrossRef]
- Gutierrez, B.L.; Macleod, N.; Edgecombe, G.D. Detecting taxonomic signal in an under-utilised character system: Geometric morphometrics of the forcipular coxae of Scutigeromorpha (Chilopoda). Zookeys 2011, 66, 49–66. [Google Scholar] [CrossRef] [Green Version]
- Perez-Gelabert, D.E.; Edgecombe, G.D. Scutigeromorph centipedes (Chilopoda: Scutigeromorpha) of the Dominican Republic, Hispaniola. Novit. Caribaea 2013, 6, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Rochow, V.B.; Müller, C.H.; Lindström, M. Spectral sensitivity of the eye of Scutigera coleoptrata (Linnaeus, 1758)(Chilopoda: Scutigeromorpha: Scutigeridae). Appl. Entomol. Zool. 2006, 41, 117–122. [Google Scholar] [CrossRef]
- Müller, C.G.; Rosenberg, J.; Richter, S.; Meyer-Rochow, V.B. The compound eye of Scutigera coleoptrata (Linnaeus, 1758) (Chilopoda: Notostigmophora): An ultrastructural reinvestigation that adds support to the Mandibulata concept. Zoomorphology 2003, 122, 191–209. [Google Scholar] [CrossRef]
- Koch, M.; Edgecombe, G.D. Peristomatic structures in Scutigeromorpha (Chilopoda): A comparative study, with new characters for higher-level systematics. Zoomorphology 2006, 125, 187–207. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Barrow, L. A new genus of Scutigerid centipedes (Chilopoda) from Western Australia, with new characters for morphological phylogenetics of Scutigeromorpha. Zootaxa 2007, 1409, 23–50. [Google Scholar] [CrossRef]
- Edgecombe, G.D.; Giribet, G. Phylogenetics of Scutigeromorph centipedes (Myriapoda: Chilopoda) with implications for species delimitation and historical biogeography of the Australian and New Caledonian faunas. Cladistics 2009, 25, 406–427. [Google Scholar] [CrossRef]
- Reeves, W.K. Discovery of an exotic population of Thereuonema tuberculata (Chilopoda: Scutigeromorpha), the Japanese House Centipede, in Ohio, U.S.A. Am. Midl. Nat. 2017, 177, 162–164. [Google Scholar] [CrossRef]
- Matsui, A.; Yahata, K. Comparative study of autotomic structures in centipedes (Arthropoda: Chilopoda). Proc. Arthropod. Embryol. Soc. Jpn 2012, 47, 11–19. [Google Scholar]
- Barber, A.D. Thereuonema tuberculata, a Scutigeromorph centipede from China, found in a warehouse at Swindon. Bull. Br. Myriap. Isopod Group 2011, 25, 49. [Google Scholar]
- Würmli, M. Synopsis der neotropischen Pselliodidae (Chilopoda: Scutigeromorpha). Stud. Neotrop. Fauna Environ. 1978, 13, 135–142. [Google Scholar] [CrossRef]
- Würmli, M. Zur Systematik der Gattung Scutigera (Chilopoda: Scutigeridae). Abh. Verh. 1977, 20, 123–131. [Google Scholar]
- Würmli, M.; Camatini, M. Taxonomic Problems in the Genus Thereuopoda (Chilopoda Scutigeromorpha: Scutigeridae): The Role of Postmaturational Moultings. In Myriapod Biology; Academic Press: London, UK, 1979; pp. 39–48. [Google Scholar]
- Würmli, M. Revision der afrikanischer Pselliodiden (Chilopoda: Scutigerida: Pselliodidae). Schubartiana 2005, 1, 3–8. [Google Scholar]
- Würmli, M. Zur Systematik der Scutigeriden Europas und Kleinasiens (Chilopoda: Scutigeromorpha). Ann. Des Nat. Mus. Wien 1973, 77, 399–408. [Google Scholar]
- Würmli, M. Die Scutigeromorpha (Chilopoda) von Costa Rica. Ueber Dendrothereua arborum Verhoeff. Stud. Neotrop. Fauna Environ. 1973, 8, 75–80. [Google Scholar] [CrossRef]
- Chagas, A., Jr.; Chaparro, E.; Jiménez, S.; Triana, H.D.D.E.; Seoane, J.C. The centipedes (Arthropoda, Myriapoda, Chilopoda) from Colombia: Part I. Scutigeromorpha and Scolopendromorpha. Zootaxa 2014, 3779, 133–156. [Google Scholar] [CrossRef] [Green Version]
- Negrisolo, E.; Minelli, A.; Valle, G. Extensive gene order rearrangement in the mitochondrial genome of the centipede Scutigera coleoptrata. J. Mol. Evol. 2004, 58, 413–423. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Boore, J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006, 21, 439–446. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Xu, K.K.; Chen, Q.P.; Ayivi, S.P.G.; Guan, J.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Three complete mitochondrial genomes of Orestes guangxiensis, Peruphasma schultei, and Phryganistria guangxiensis (Insecta: Phasmatodea) and their phylogeny. Insects 2021, 12, 779. [Google Scholar] [CrossRef] [PubMed]
- Ayivi, S.P.G.; Tong, Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The mitochondrial genomes of 18 new Pleurosticti (Coleoptera: Scarabaeidae) exhibit a novel trnQ-NCR-trnI-trnM gene rearrangement and clarify phylogenetic relationships of subfamilies within Scarabaeidae. Insects 2021, 12, 1025. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.Y.; Shen, S.Q.; Zhang, Z.Y.; Xu, X.D.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Comparative mitogenomes of two coreamachilis species (Microcoryphia: Machilidae) along with phylogenetic analyses of Microcoryphia. Insects 2021, 12, 795. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Wu, L.; Ayivi, S.P.G.; Storey, K.B.; Ma, Y.; Yu, D.N.; Zhang, J.Y. Cryptic species exist in Vietnamella sinensis Hsu, 1936 (Insecta: Ephemeroptera) from studies of complete mitochondrial genomes. Insects 2022, 13, 412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Guan, J.Y.; Cao, Y.R.; Dai, X.Y.; Yu, D.n.; Zhang, J.Y. Mitogenome analysis of four Lamiinae species (Coleoptera: Cerambycidae) and gene expression responses by monochamus alternatus when Infected with the Parasitic Nematode, Bursaphelenchus mucronatus. Insects 2021, 12, 453. [Google Scholar] [CrossRef]
- Ye, S.P.; Huang, H.; Zheng, R.Q.; Zhang, J.Y.; Yang, G.; Xu, S.X. Phylogeographic analyses strongly suggest cryptic speciation in the giant spiny frog (Dicroglossidae: Paa spinosa) and interspecies hybridization in Paa. PLoS ONE 2013, 8, e70403. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.N.; Zhang, J.Y.; Li, P.; Zheng, R.Q.; Shao, C. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the cyt b gene and the complete mt genome. PLoS ONE 2015, 10, e0124825. [Google Scholar] [CrossRef]
- Xu, X.D.; Guan, J.Y.; Zhang, Z.Y.; Cao, Y.R.; Cai, Y.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects 2021, 12, 656. [Google Scholar] [CrossRef]
- Jia, W.Z.; Yan, H.B.; Lou, Z.Z.; Ni, X.W.; Dyachenko, V.; Li, H.M.; Littlewood, D.T.J. Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Trop. 2012, 123, 154–163. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhou, C.F.; Gai, Y.H.; Song, D.X.; Zhou, K.Y. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene 2008, 424, 18–24. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000 Res. 2019, 8, 1874. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, D.; Patrick, M.; Guillaume, S. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef] [Green Version]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Gao, F.l.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Negrisolo, E.; Minelli, A.; Valle, G. The mitochondrial genome of the house centipede Scutigera and the monophyly versus paraphyly of Myriapods. Mol. Biol. Evol. 2004, 21, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.E.; Lapraz, F.; Rhodes, A.C.; Telford, M.J. The complete mitochondrial genome of the Geophilomorph centipede Strigamia maritima. PLoS ONE 2015, 10, e0121369. [Google Scholar] [CrossRef] [Green Version]
- Gai, Y.H.; Ma, H.Q.; Ma, J.Y.; Li, C.X.; Yang, Q. The complete mitochondrial genome of Scolopocryptops sp. (Chilopoda: Scolopendromorpha: Scolopocryptopidae). Mitochondrial DNA 2014, 25, 192–193. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.Y.; Wang, S.B.; Huang, B.S.; Liu, H.G.; Liu, Y.F. The complete mitochondrial genome sequence of Scolopendra mutilans L. Koch, 1878 (Scolopendromorpha, Scolopendridae), with a comparative analysis of other centipede genomes. Zookeys 2020, 925, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Hwang, U.W.; Friedrich, M.; Tautz, D.; Park, C.J.; Kim, W. Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature 2001, 413, 154–157. [Google Scholar] [CrossRef]
- Gai, Y.H.; Ma, H.Q.; Sun, X.Y.; Ma, J.Y.; Li, C.X.; Yang, Q. The complete mitochondrial genome of Cermatobius longicornis (Chilopoda: Lithobiomorpha: Henicopidae). Mitochondrial DNA 2013, 24, 331–332. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.J.; Nguyen, A.D.; Jang, K.H.; Choi, E.H.; Ryu, S.H.; Hwang, U.W. The complete mitochondrial genome of the Korean endemic millipede Anaulaciulus koreanus (Verhoeff, 1937), with notes on the gene arrangement of millipede orders. Zootaxa 2017, 4329, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Welch, J.J.; Suchard, M.A. Phylogeography takes a relaxed random walk in continuous space and time. Mol. Biol. Evol. 2010, 27, 1877–1885. [Google Scholar] [CrossRef] [Green Version]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Hanada, T.; Suzuki, T.; Yokogawa, T.; Takemoto-Hori, C.; Sprinzl, M.; Watana Be, K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells 2001, 6, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H. Surprising contribution to aminoacylation and translation of non-Watson–Crick pairs in tRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4570–4575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, D.A. Replication of animal mitochondrial DNA. Cell 1982, 28, 693–705. [Google Scholar] [CrossRef]
- Wesener, T.; Voigtländer, K.; Decker, P.; Oeyen, J.P.; Spelda, J. Barcoding of central European Cryptops centipedes reveals large interspecific distances with ghost lineages and new species records from Germany and Austria (Chilopoda, Scolopendromorpha). Zookeys 2016, 564, 21–46. [Google Scholar] [CrossRef] [Green Version]
- Siriwut, W.; Edgecombe, G.D.; Sutcharit, C.; Tongkerd, P.; Panha, S. First record of the African-Indian centipede genus Digitipes Attems, 1930 (Scolopendromorpha: Otostigminae) from Myanmar, and the systematic position of a new species based on molecular phylogenetics. Zootaxa 2015, 3931, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Oeyen, J.P.; Funke, S.; Boehme, W.; Wesener, T. The evolutionary history of the rediscovered Austrian population of the giant centipede Scolopendra cingulata Latreille 1829 (Chilopoda, Scolopendromorpha). PLoS ONE 2014, 9, e108650. [Google Scholar] [CrossRef] [Green Version]
- Dohle, W. Phylogenetic pathways in the Chilopoda. Bijdr. Tot Dierkd. 1985, 55, 55–56. [Google Scholar]
- Shear, W.A.; Bonamo, P.M. Devonobiomorpha, a new order of centipeds (Chilopoda) from the Middle Devonian of Gilboa, New York State, USA, and the phylogeny of centiped orders. Am. Mus. Novit. 1988, 2927, 1–30. [Google Scholar]



| Species | Specimen No. | Sampling Localities | Accession Number |
|---|---|---|---|
| Thereuonema tuberculata | HNYY01 | Nanyang, Henan, China | OK513221 |
| JXYY02 | Nanchang, Jiangxi, China | OL449685 | |
| SDYY03 | Jinan, Shandong, China | ON058988 | |
| YNYY11 | Dali, Yunnan, China | ON058989 |
| Class | Order | Family | Genus | Species | Length (bp) | GenBank No. | References |
|---|---|---|---|---|---|---|---|
| Diplopoda | Helminthomorpha | Julidae | Anaulaciulus | Anaulaciulus koreanus | 14,916 | KX096886 | [63] |
| Spirobolidae | Spirobolus | Spirobolus bungii | 14,879 | MT767838 | Direct Submission | ||
| Chilopoda | Scolopendromorpha | Cryptopidae | Scolopocryptops | Scolopocryptops sp. 1 YG-2013 | 15,119 | KC200076 | [58] |
| Scolopendridae | Scolopendra | Scolopendra mutilans | 15,011 | MN317390 | [59] | ||
| Scolopendra mutilans | 15,030 | MT175377 | Unpublished | ||||
| Scolopendra subspinipes | 14,637 | MN642577 | Unpublished | ||||
| Scolopendra dehaani | 14,538 | KY947341 | Unpublished | ||||
| Lithobiomorpha | Henicopidae | Cermatobius | Cermatobius longicornis | 16,833 | KC155628 | [61] | |
| Ethopolyidae | Bothropolys | Bothropolys sp. SP-2004 | 15,139 | AY691655 | Direct Submission | ||
| Lithobiidae | Lithobius | Lithobius forficatus | 15,437 | AJ270997 | [60] | ||
| Lithobius forficatus | 15,695 | AF309492 | [62] | ||||
| Lithobius forficatus | 15,038 | MT862427 | Unpublished | ||||
| Geophilomorpha | Mecistocephalidae | Mecistocephalus | Mecistocephalus marmoratus | 15,279 | KX774322 | Unpublished | |
| Linotaeniidae | Strigamia | Strigamia maritima | 14,983 | KP173664 | [57] | ||
| Scutigeromorpha | Scutigeridae | Scutigera | Scutigera coleoptrata | 14,922 | AJ507061 | [56] | |
| Thereuonema | Thereuonema tuberculata NY | 14,905 | OK513221 | This study | |||
| Thereuonema tuberculata NC | 14,906 | OL449685 | This study | ||||
| Thereuonema tuberculata JN | 14,909 | ON058988 | This study | ||||
| Thereuonema tuberculata DL | 14,903 | ON058989 | This study |
| Region | T. tuberculata NY | T. tuberculata NC | T. tuberculata JN | T. tuberculata DL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | A + T(%) | AT Skew | GC Skew | Length (bp) | A + T(%) | AT Skew | GC Skew | Length (bp) | A + T(%) | AT Skew | GC Skew | Length (bp) | A + T(%) | AT Skew | GC Skew | |
| Mito | 14,905 | 71.8 | 0.005 | −0.287 | 14,906 | 71.9 | 0.013 | −0.292 | 14909 | 71.7 | 0.018 | −0.303 | 14,903 | 71.0 | 0.01 | −0.273 |
| PCGs | 11,079 | 71.1 | 0.001 | −0.287 | 11,082 | 70.9 | 0.010 | −0.288 | 11082 | 70.8 | 0.017 | −0.305 | 11,088 | 70.0 | 0.009 | −0.273 |
| rRNAs | 1950 | 73.2 | −0.028 | 0.368 | 1954 | 73.1 | −0.039 | 0.392 | 1956 | 72.8 | −0.03 | 0.375 | 1971 | 73.1 | −0.022 | 0.343 |
| tRNAs | 1376 | 73.3 | 0.010 | −0.196 | 1374 | 73.7 | 0.019 | −0.190 | 1374 | 73.0 | 0.034 | −0.206 | 1377 | 72.7 | 0.026 | −0.184 |
| A + T-rich region | 461 | 80.3 | 0.022 | −0.143 | 463 | 83.4 | 0.005 | −0.143 | 463 | 82.5 | 0.005 | −0.086 | 439 | 81.5 | 0.006 | −0.136 |
| Sample | Complete Mitochondrial Genomes/Partial COX1 Genes | |||
|---|---|---|---|---|
| T. tuberculata NY | T. tuberculata NC | T. tuberculata JN | T. tuberculata DL | |
| T. tuberculata NY | ||||
| T. tuberculata NC | 0.097/0.103 | |||
| T. tuberculata JN | 0.100/0.116 | 0.077/0.073 | ||
| T. tuberculata DL | 0.152/0.145 | 0.150/0.140 | 0.151/0.135 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-M.; Zhang, L.-H.; Lin, Y.-J.; Zheng, Y.-M.; Jin, W.-T.; Storey, K.B.; Yu, D.-N.; Zhang, J.-Y. The Genetic Diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the Phylogenetic Relationship of Scutigeromorpha Using the Mitochondrial Genome. Insects 2022, 13, 620. https://doi.org/10.3390/insects13070620
Yang Y-M, Zhang L-H, Lin Y-J, Zheng Y-M, Jin W-T, Storey KB, Yu D-N, Zhang J-Y. The Genetic Diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the Phylogenetic Relationship of Scutigeromorpha Using the Mitochondrial Genome. Insects. 2022; 13(7):620. https://doi.org/10.3390/insects13070620
Chicago/Turabian StyleYang, Yong-Mei, Li-Hua Zhang, Yi-Jie Lin, Yi-Meng Zheng, Wan-Ting Jin, Kenneth B. Storey, Dan-Na Yu, and Jia-Yong Zhang. 2022. "The Genetic Diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the Phylogenetic Relationship of Scutigeromorpha Using the Mitochondrial Genome" Insects 13, no. 7: 620. https://doi.org/10.3390/insects13070620






