Large-Scale Monitoring of the Frequency of Ryanodine Receptor Target-Site Mutations Conferring Diamide Resistance in Brazilian Field Populations of Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fall Armyworm (FAW) Strains
2.2. Phenotypic Resistance Monitoring of Faw Using a Diagnostic Concentration of Flubendiamide
2.3. F2 Screen for Flubendiamide Resistance Alleles
2.4. Genetics and Inheritance of S. frugiperda Resistance to Flubendiamide
2.4.1. Dominance of Resistance
2.4.2. Number of Genes Associated with Resistance
2.5. Cross-Resistance of Flubendiamide Resistance to Anthranilic Diamides
2.6. Partial Sequencing of the Ryanodine Receptor (RyR) C-Terminal Transmembrane Domain
2.7. PCR-Based Allelic Discrimination Assay for Genotyping
3. Results
3.1. Phenotypic Resistance Monitoring of FAW Using a Diagnostic Concentration of Flubendiamide
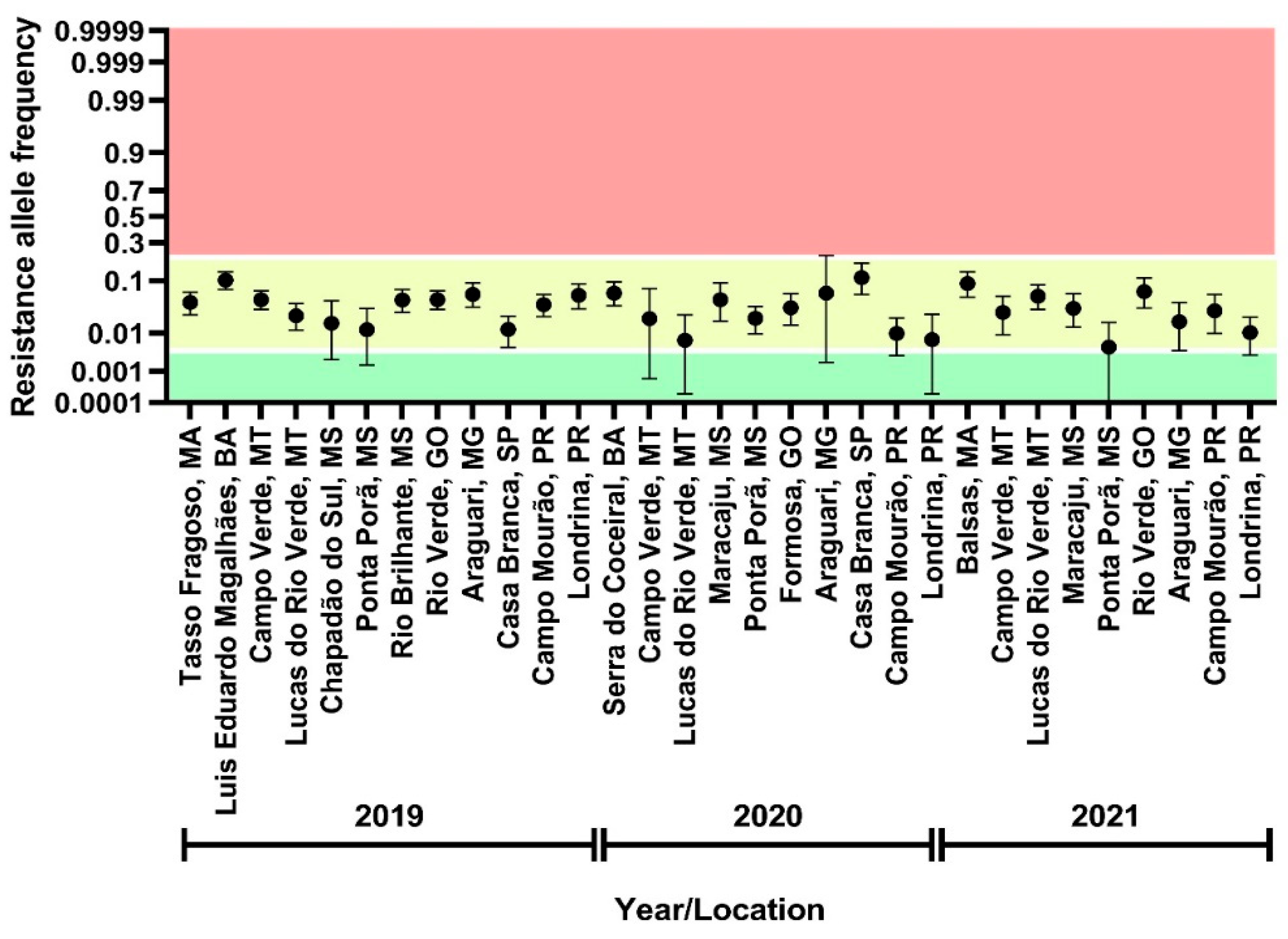
3.2. Assessment of Flubendiamide Resistance Allele Frequency
3.3. Establishment of Flubendiamide Resistant FAW Strains BA-R and TF-R
3.4. Genetics of Resistance in FAW Strains BA-R and TF-R
3.5. Cross-Resistance of FAW Strains BA-R and TF-R to Anthranilic DIamides
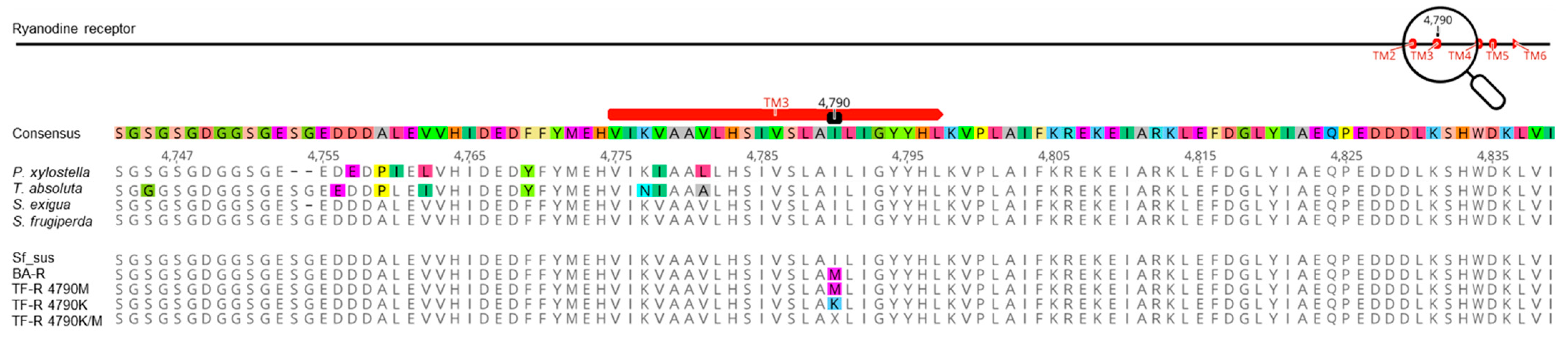
3.6. Ryanodine Receptor (RyR) C-Terminal Transmembrane Sequencing
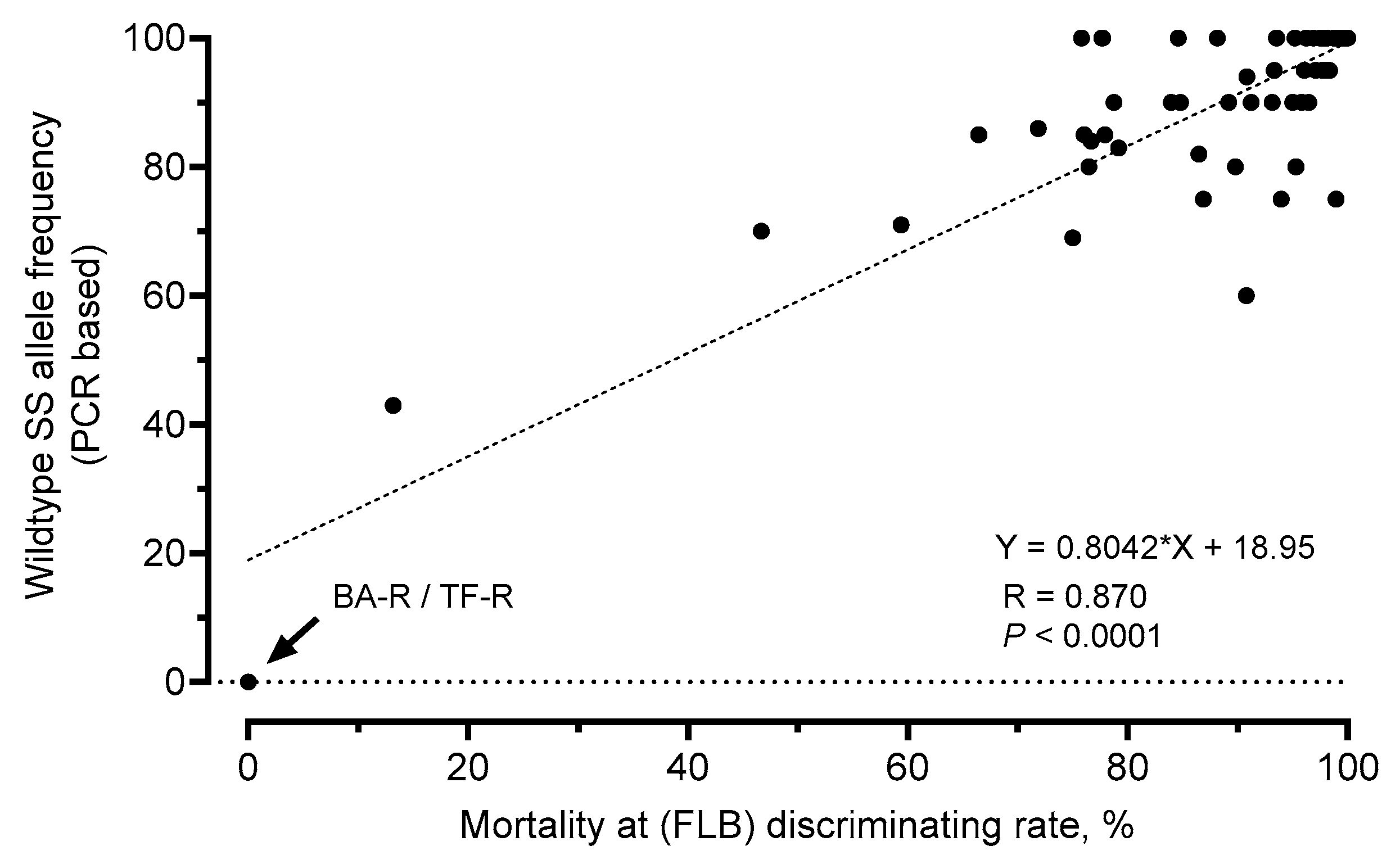
3.7. Genotyping of FAW Field Samples by a PCR-Based Allelic Discrimination Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overton, K.; Maino, J.L.; Day, R.; Umina, P.A.; Bett, B.; Carnovale, D.; Ekesi, S.; Meagher, R.; Reynolds, O.L. Global Crop Impacts, Yield Losses and Action Thresholds for Fall Armyworm (Spodoptera frugiperda): A Review. Crop Prot. 2021, 145, 105641. [Google Scholar] [CrossRef]
- Sparks, A.N. A Review of the Biology of the Fall Armyworm. Fla. Entomol. 1979, 62, 82–87. [Google Scholar] [CrossRef]
- Barros, E.M.; Torres, J.B.; Ruberson, J.R.; Oliveira, M.D. Development of Spodoptera frugiperda on Different Hosts and Damage to Reproductive Structures in Cotton. Entomol. Exp. Appl. 2010, 137, 237–245. [Google Scholar] [CrossRef]
- Montezano, D.G.; Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula-Moraes, S.V.; Peterson, J.A.; Hunt, T.E. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, R.J.; Dourado, P.M.; Berger, G.U.; de S. Fernandes, D.; Omoto, C.; Willse, A.; Martinelli, S.; Head, G.P.; Corrêa, A.S. Large-Scale Assessment of Lepidopteran Soybean Pests and Efficacy of Cry1Ac Soybean in Brazil. Sci. Rep. 2021, 11, 15956. [Google Scholar] [CrossRef] [PubMed]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First Report of Outbreaks of the Fall Armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [Green Version]
- Niassy, S.; Agbodzavu, M.K.; Kimathi, E.; Mutune, B.; Abdel-Rahman, E.F.M.; Salifu, D.; Hailu, G.; Belayneh, Y.T.; Felege, E.; Tonnang, H.E.Z.; et al. Bioecology of Fall Armyworm Spodoptera frugiperda (J. E. Smith), Its Management and Potential Patterns of Seasonal Spread in Africa. PLoS ONE 2021, 16, e0249042. [Google Scholar] [CrossRef]
- Sharanabasappa; Kalleshwaraswamy, C.M.; Asokan, R.; Swamy, H.M.M.; Maruthi, M.S.; Pavithra, H.B.; Hegbe, K.; Navi, S.; Prabhu, S.T.; Goergen, G.E. First Report of the Fall Armyworm, Spodoptera frugiperda (J E Smith) (Lepidoptera: Noctuidae), an Alien Invasive Pest on Maize in India. Pest Manag. Hortic. Ecosyst. 2018, 24, 23–29. [Google Scholar]
- Maino, J.L.; Schouten, R.; Overton, K.; Day, R.; Ekesi, S.; Bett, B.; Barton, M.; Gregg, P.C.; Umina, P.A.; Reynolds, O.L. Regional and Seasonal Activity Predictions for Fall Armyworm in Australia. Curr. Res. Insect Sci. 2021, 1, 100010. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Q.; Zhang, H.; Wu, K. Spread of Invasive Migratory Pest Spodoptera frugiperda and Management Practices throughout China. J. Integr. Agric. 2021, 20, 637–645. [Google Scholar] [CrossRef]
- Burtet, L.M.; Bernardi, O.; Melo, A.A.; Pes, M.P.; Strahl, T.T.; Guedes, J.V. Managing Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), with Bt Maize and Insecticides in Southern Brazil. Pest Manag. Sci. 2017, 73, 2569–2577. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Sánchez, F.A.; Rivera, G.; Bocanegra-García, V.; Martínez-Padrón, H.Y.; Berrones-Morales, M.; Niño-García, N.; Herrera-Mayorga, V. Advances in Control Strategies against Spodoptera frugiperda. A Review. Molecules 2021, 26, 5587. [Google Scholar] [CrossRef] [PubMed]
- Diez-Rodríguez, G.I.; Omoto, C. Herança da resistência de Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) a lambda-cialotrina. Neotrop. Entomol. 2001, 30, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, R.A.; Omoto, C.; Field, L.M.; Williamson, M.S.; Bass, C. Investigating the Molecular Mechanisms of Organophosphate and Pyrethroid Resistance in the Fall Armyworm Spodoptera frugiperda. PLoS ONE 2013, 8, e62268. [Google Scholar] [CrossRef] [Green Version]
- Do Nascimento, A.R.B.; Farias, J.R.; Bernardi, D.; Horikoshi, R.J.; Omoto, C. Genetic Basis of Spodoptera frugiperda (Lepidoptera: Noctuidae) Resistance to the Chitin Synthesis Inhibitor Lufenuron. Pest Manag. Sci. 2016, 72, 810–815. [Google Scholar] [CrossRef]
- Okuma, D.M.; Bernardi, D.; Horikoshi, R.J.; Bernardi, O.; Silva, A.P.; Omoto, C. Inheritance and Fitness Costs of Spodoptera frugiperda (Lepidoptera: Noctuidae) Resistance to Spinosad in Brazil. Pest Manag. Sci. 2018, 74, 1441–1448. [Google Scholar] [CrossRef]
- Boaventura, D.; Bolzan, A.; Padovez, F.E.; Okuma, D.M.; Omoto, C.; Nauen, R. Detection of a Ryanodine Receptor Target-Site Mutation in Diamide Insecticide Resistant Fall Armyworm, Spodoptera frugiperda. Pest Manag. Sci. 2020, 76, 47–54. [Google Scholar] [CrossRef]
- Bolzan, A.; Padovez, F.E.; Nascimento, A.R.; Kaiser, I.S.; Lira, E.C.; Amaral, F.S.; Kanno, R.H.; Malaquias, J.B.; Omoto, C. Selection and Characterization of the Inheritance of Resistance of Spodoptera frugiperda (Lepidoptera: Noctuidae) to Chlorantraniliprole and Cross-Resistance to Other Diamide Insecticides. Pest Manag. Sci. 2019, 75, 2682–2689. [Google Scholar] [CrossRef]
- Muraro, D.S.; de Oliveira Abbade Neto, D.; Kanno, R.H.; Kaiser, I.S.; Bernardi, O.; Omoto, C. Inheritance Patterns, Cross-Resistance and Synergism in Spodoptera frugiperda (Lepidoptera: Noctuidae) Resistant to Emamectin Benzoate. Pest Manag. Sci. 2021, 77, 5049–5057. [Google Scholar] [CrossRef]
- Omoto, C.; Bernardi, O.; Salmeron, E.; Sorgatto, R.J.; Dourado, P.M.; Crivellari, A.; Carvalho, R.A.; Willse, A.; Martinelli, S.; Head, G.P. Field-Evolved Resistance to Cry1Ab Maize by Spodoptera frugiperda in Brazil. Pest Manag. Sci. 2016, 72, 1727–1736. [Google Scholar] [CrossRef]
- Farias, J.R.; Andow, D.A.; Horikoshi, R.J.; Sorgatto, R.J.; Fresia, P.; dos Santos, A.C.; Omoto, C. Field-Evolved Resistance to Cry1F Maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 2014, 64, 150–158. [Google Scholar] [CrossRef]
- Machado, E.; dos S. Rodrigues, G.L., Jr.; Führ, F.M.; Zago, S.; Marques, L.; Santos, A.; Nowatzki, T.; Dahmer, M.; Omoto, C.; Bernardi, O. Cross-Crop Resistance of Spodoptera frugiperda Selected on Bt Maize to Genetically-Modified Soybean Expressing Cry1Ac and Cry1F Proteins in Brazil. Sci. Rep. 2020, 10, 10080. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, D.; Salmeron, E.; Horikoshi, R.J.; Bernardi, O.; Dourado, P.M.; Carvalho, R.A.; Martinelli, S.; Head, G.P.; Omoto, C. Cross-Resistance between Cry1 Proteins in Fall Armyworm (Spodoptera frugiperda) May Affect the Durability of Current Pyramided Bt Maize Hybrids in Brazil. PLoS ONE 2015, 10, e0140130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardi, O.; Bernardi, D.; Horikoshi, R.J.; Okuma, D.M.; Miraldo, L.L.; Fatoretto, J.; Medeiros, F.C.; Burd, T.; Omoto, C. Selection and Characterization of Resistance to the Vip3Aa20 Protein from Bacillus Thuringiensis in Spodoptera frugiperda. Pest Manag. Sci. 2016, 72, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Nauen, R. Insecticide Mode of Action: Return of the Ryanodine Receptor. Pest Manag. Sci. 2006, 62, 690–692. [Google Scholar] [CrossRef] [PubMed]
- Sattelle, D.B.; Cordova, D.; Cheek, T.R. Insect Ryanodine Receptors: Molecular Targets for Novel Pest Control Chemicals. Invert. Neurosci. 2008, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Jeanguenat, A. The Story of a New Insecticidal Chemistry Class: The Diamides. Pest Manag. Sci. 2013, 69, 7–14. [Google Scholar] [CrossRef]
- Tohnishi, M.; Nakao, H.; Furuya, T.; Seo, A.; Kodama, H.; Tsubata, K.; Fujioka, S.; Kodama, H.; Hirooka, T.; Nishimatsu, T. Flubendiamide, a Novel Insecticide Highly Active against Lepidopterous Insect Pests. J. Pestic. Sci. 2005, 30, 354–360. [Google Scholar] [CrossRef] [Green Version]
- Spark2 Spark Smarter Decisions, Valinhos, SP. Available online: http://spark-ie.com.br (accessed on 1 June 2022).
- Cordova, D.; Benner, E.A.; Sacher, M.D.; Rauh, J.J.; Sopa, J.S.; Lahm, G.P.; Selby, T.P.; Stevenson, T.M.; Flexner, L.; Gutteridge, S.; et al. Anthranilic Diamides: A New Class of Insecticides with a Novel Mode of Action, Ryanodine Receptor Activation. Pestic. Biochem. Physiol. 2006, 84, 196–214. [Google Scholar] [CrossRef]
- Lahm, G.P.; Stevenson, T.M.; Selby, T.P.; Freudenberger, J.H.; Cordova, D.; Flexner, L.; Bellin, C.A.; Dubas, C.M.; Smith, B.K.; Hughes, K.A.; et al. RynaxypyrTM: A New Insecticidal Anthranilic Diamide That Acts as a Potent and Selective Ryanodine Receptor Activator. Bioorg. Med. Chem. Lett. 2007, 17, 6274–6279. [Google Scholar] [CrossRef]
- Lahm, G.P.; Selby, T.P.; Freudenberger, J.H.; Stevenson, T.M.; Myers, B.J.; Seburyamo, G.; Smith, B.K.; Flexner, L.; Clark, C.E.; Cordova, D. Insecticidal Anthranilic Diamides: A New Class of Potent Ryanodine Receptor Activators. Bioorg. Med. Chem. Lett. 2005, 15, 4898–4906. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Kiyonaka, S.; Sawaguchi, Y.; Tohnishi, M.; Masaki, T.; Yasokawa, N.; Mizuno, Y.; Mori, E.; Inoue, K.; Hamachi, I.; et al. Molecular Characterization of Flubendiamide Sensitivity in the Lepidopterous Ryanodine Receptor Ca2+ Release Channel. Biochemistry 2009, 48, 10342–10352. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Haji-Ghassemi, O.; Ma, D.; Jiang, H.; Lin, L.; Yao, L.; Samurkas, A.; Li, Y.; Wang, Y.; Cao, P.; et al. Structural Basis for Diamide Modulation of Ryanodine Receptor. Nat. Chem. Biol. 2020, 16, 1246–1254. [Google Scholar] [CrossRef]
- Qi, S.; Lümmen, P.; Nauen, R.; Casida, J.E. Diamide Insecticide Target Site Specificity in the Heliothis and Musca Ryanodine Receptors Relative to Toxicity. J. Agric. Food Chem. 2014, 62, 4077–4082. [Google Scholar] [CrossRef] [PubMed]
- Troczka, B.; Zimmer, C.T.; Elias, J.; Schorn, C.; Bass, C.; Davies, T.G.E.; Field, L.M.; Williamson, M.S.; Slater, R.; Nauen, R. Resistance to Diamide Insecticides in Diamondback Moth, Plutella xylostella (Lepidoptera: Plutellidae) Is Associated with a Mutation in the Membrane-Spanning Domain of the Ryanodine Receptor. Insect Biochem. Mol. Biol. 2012, 42, 873–880. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Y. High Levels of Resistance to Chlorantraniliprole Evolved in Field Populations of Plutella xylostella. J. Econ. Entomol. 2012, 105, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Richardson, E.B.; Troczka, B.J.; Gutbrod, O.; Davies, T.G.E.; Nauen, R. Diamide Resistance: 10 Years of Lessons from Lepidopteran Pests. J. Pest Sci. 2020, 93, 911–928. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Moreno, R.; Mota-Sanchez, D.; Blanco, C.A.; Whalon, M.E.; Terán-Santofimio, H.; Rodriguez-Maciel, J.C.; DiFonzo, C. Field-Evolved Resistance of the Fall Armyworm (Lepidoptera: Noctuidae) to Synthetic Insecticides in Puerto Rico and Mexico. J. Econ. Entomol. 2019, 112, 792–802. [Google Scholar] [CrossRef]
- Guo, L.; Liang, P.; Zhou, X.; Gao, X. Novel Mutations and Mutation Combinations of Ryanodine Receptor in a Chlorantraniliprole Resistant Population of Plutella xylostella (L.). Sci. Rep. 2014, 4, 6924. [Google Scholar] [CrossRef] [Green Version]
- Steinbach, D.; Gutbrod, O.; Lümmen, P.; Matthiesen, S.; Schorn, C.; Nauen, R. Geographic Spread, Genetics and Functional Characteristics of Ryanodine Receptor Based Target-Site Resistance to Diamide Insecticides in Diamondback Moth, Plutella xylostella. Insect Biochem. Mol. Biol. 2015, 63, 14–22. [Google Scholar] [CrossRef]
- Roditakis, E.; Steinbach, D.; Moritz, G.; Vasakis, E.; Stavrakaki, M.; Ilias, A.; García-Vidal, L.; del R. Martínez-Aguirre, M.; Bielza, P.; Morou, E.; et al. Ryanodine Receptor Point Mutations Confer Diamide Insecticide Resistance in Tomato Leafminer, Tuta absoluta (Lepidoptera: Gelechiidae). Insect Biochem. Mol. Biol. 2017, 80, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, L.; Chen, Q.; Qin, W.; Huang, S.; Jiang, Y.; Qin, H. Chlorantraniliprole Resistance and Its Biochemical and New Molecular Target Mechanisms in Laboratory and Field Strains of Chilo suppressalis (Walker). Pest Manag. Sci. 2018, 74, 1416–1423. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Zhao, D.-D.; Zhang, S.; Zhou, L.-Q.; Wang, X.; Gao, C.-F.; Wu, S.-F. Monitoring and Mechanisms of Insecticide Resistance in Chilo suppressalis (Lepidoptera: Crambidae), with Special Reference to Diamides. Pest Manag. Sci. 2017, 73, 1169–1178. [Google Scholar] [CrossRef]
- Kim, J.; Nam, H.Y.; Kwon, M.; Choi, J.H.; Cho, S.R.; Kim, G.-H. Novel Diamide Resistance-Linked Mutation in Korean Spodoptera Exigua and a LAMP Assay Based on a Mutation-Associated Intronic InDel. J. Pest Sci. 2021, 94, 1017–1029. [Google Scholar] [CrossRef]
- Zuo, Y.-Y.; Ma, H.-H.; Lu, W.-J.; Wang, X.-L.; Wu, S.-W.; Nauen, R.; Wu, Y.-D.; Yang, Y.-H. Identification of the Ryanodine Receptor Mutation I4743M and Its Contribution to Diamide Insecticide Resistance in Spodoptera exigua (Lepidoptera: Noctuidae). Insect Sci. 2020, 27, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Richardson, E.; Homem, R.A.; Troczka, B.J.; George, C.H.; Ebbinghaus-Kintscher, U.; Williamson, M.S.; Nauen, R.; Davies, T.E. Diamide Insecticide Resistance in Transgenic Drosophila and Sf9-Cells Expressing a Full-Length Diamondback Moth Ryanodine Receptor Carrying an I4790M Mutation. Pest Manag. Sci. 2022, 78, 869–880. [Google Scholar] [CrossRef]
- Troczka, B.J.; Williams, A.J.; Williamson, M.S.; Field, L.M.; Lüemmen, P.; Davies, T.G.E. Stable Expression and Functional Characterisation of the Diamondback Moth Ryanodine Receptor G4946E Variant Conferring Resistance to Diamide Insecticides. Sci. Rep. 2015, 5, 14680. [Google Scholar] [CrossRef]
- Douris, V.; Papapostolou, K.-M.; Ilias, A.; Roditakis, E.; Kounadi, S.; Riga, M.; Nauen, R.; Vontas, J. Investigation of the Contribution of RyR Target-Site Mutations in Diamide Resistance by CRISPR/Cas9 Genome Modification in Drosophila. Insect Biochem. Mol. Biol. 2017, 87, 127–135. [Google Scholar] [CrossRef]
- Zuo, Y.; Wang, H.; Xu, Y.; Huang, J.; Wu, S.; Wu, Y.; Yang, Y. CRISPR/Cas9 Mediated G4946E Substitution in the Ryanodine Receptor of Spodoptera Exigua Confers High Levels of Resistance to Diamide Insecticides. Insect Biochem. Mol. Biol. 2017, 89, 79–85. [Google Scholar] [CrossRef]
- Teixeira, L.A.; Andaloro, J.T. Diamide Insecticides: Global Efforts to Address Insect Resistance Stewardship Challenges. Pestic. Biochem. Physiol. 2013, 106, 76–78. [Google Scholar] [CrossRef]
- Andow, D.A.; Alstad, D.N. F2 Screen for Rare Resistance Alleles. J. Econ. Entomol. 1998, 91, 572–578. [Google Scholar] [CrossRef]
- Andow, D.; Alstad, D.N. Letter to the Editor. J. Econ. Entomol. 1999, 92, 755–758. [Google Scholar] [CrossRef]
- European Environment Agency. R Core Team. 2020. Available online: https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (accessed on 9 June 2022).
- Da Silva Ribeiro, R. Monitoramento da Suscetibilidade de Populações de Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) a Inseticidas Diamidas no Brasil. Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brasil, 2014. [Google Scholar]
- Bourguet, D.; Genissel, A.; Raymond, M. Insecticide Resistance and Dominance Levels. J. Econ. Entomol. 2000, 93, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.F. A Formula for Determining Degree of Dominance in Cases of Monofactorial Inheritance of Resistance to Chemicals. Bull. World Health Organ. 1968, 38, 325–326. [Google Scholar]
- Roush, R.T.; Daly, J.C. The Role of Population Genetics in Resistance Research and Management. In Pesticide Resistance in Arthropods; Roush, R.T., Tabashnik, B.E., Eds.; Springer: Boston, MA, USA, 1990; pp. 97–152. ISBN 978-1-4684-6429-0. [Google Scholar]
- Tsukamoto, M. Methods of Genetic Analysis of Insecticide Resistance. In Pest Resistance to Pesticides; Georghiou, G.P., Saito, T., Eds.; Springer: Boston, MA, USA, 1983; pp. 71–98. ISBN 978-1-4684-4466-7. [Google Scholar]
- Lande, R. The minimum number of genes contributing to quantitative variation between and within populations. Genetics 1981, 99, 541–553. [Google Scholar] [CrossRef]
- Robertson, J.L.; Russell, R.M.; Preisler, H.K.; Savin, N.E. Bioassays with Arthropods, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Cho, S.-R.; Kyung, Y.; Shin, S.; Kang, W.-J.; Jung, D.H.; Lee, S.-J.; Park, G.-H.; Kim, S.I.; Cho, S.W.; Kim, H.K.; et al. Susceptibility of Field Populations of Plutella xylostella and Spodoptera exigua to Four Diamide Insecticides. Korean J. Appl. Entomol. 2018, 57, 43–50. [Google Scholar] [CrossRef]
- Roditakis, E.; Vasakis, E.; García-Vidal, L.; del Rosario Martínez-Aguirre, M.; Rison, J.L.; Haxaire-Lutun, M.O.; Nauen, R.; Tsagkarakou, A.; Bielza, P. A Four-Year Survey on Insecticide Resistance and Likelihood of Chemical Control Failure for Tomato Leaf Miner Tuta Absoluta in the European/Asian Region. J. Pest Sci. 2018, 91, 421–435. [Google Scholar] [CrossRef]
- Uchiyama, T.; Ozawa, A. Rapid Development of Resistance to Diamide Insecticides in the Smaller Tea Tortrix, Adoxophyes honmai (Lepidoptera: Tortricidae), in the Tea Fields of Shizuoka Prefecture, Japan. Appl. Entomol. Zool. 2014, 49, 529–534. [Google Scholar] [CrossRef]
- Jouraku, A.; Kuwazaki, S.; Miyamoto, K.; Uchiyama, M.; Kurokawa, T.; Mori, E.; Mori, M.X.; Mori, Y.; Sonoda, S. Ryanodine Receptor Mutations (G4946E and I4790K) Differentially Responsible for Diamide Insecticide Resistance in Diamondback Moth, Plutella xylostella L. Insect Biochem. Mol. Biol. 2020, 118, 103308. [Google Scholar] [CrossRef]
- Sparks, T.C.; Nauen, R. IRAC: Mode of Action Classification and Insecticide Resistance Management. Pestic. Biochem. Physiol. 2015, 121, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Ward, C.M.; Perry, K.D.; Baker, G.; Powis, K.; Heckel, D.G.; Baxter, S.W. A Haploid Diamondback Moth (Plutella xylostella L.) Genome Assembly Resolves 31 Chromosomes and Identifies a Diamide Resistance Mutation. Insect Biochem. Mol. Biol. 2021, 138, 103622. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Qian, C.; Wang, D.; Wang, F.; Zhao, S.; Yang, Y.; Baxter, S.W.; Wang, X.; Wu, Y. Varying Contributions of Three Ryanodine Receptor Point Mutations to Diamide Insecticide Resistance in Plutella xylostella. Pest Manag. Sci. 2021, 77, 4874–4883. [Google Scholar] [CrossRef] [PubMed]
- Richardson, E.; Troczka, B.J.; Gutbrod, O.; Ebbinghaus-Kintscher, U.; Williamson, M.S.; George, C.H.; Nauen, R.; Davies, T.G.E. Chimeric Investigations into the Diamide Binding Site on the Lepidopteran Ryanodine Receptor. Int. J. Mol. Sci. 2021, 22, 13033. [Google Scholar] [CrossRef]
- Boaventura, D.; Martin, M.; Pozzebon, A.; Mota-Sanchez, D.; Nauen, R. Monitoring of Target-Site Mutations Conferring Insecticide Resistance in Spodoptera Frugiperda. Insects 2020, 11, 545. [Google Scholar] [CrossRef]
- Guan, F.; Zhang, J.; Shen, H.; Wang, X.; Padovan, A.; Walsh, T.K.; Tay, W.T.; Gordon, K.H.J.; James, W.; Czepak, C.; et al. Whole-Genome Sequencing to Detect Mutations Associated with Resistance to Insecticides and Bt Proteins in Spodoptera frugiperda. Insect Sci. 2021, 28, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Yainna, S.; Nègre, N.; Silvie, P.J.; Brévault, T.; Tay, W.T.; Gordon, K.; dAlençon, E.; Walsh, T.; Nam, K. Geographic Monitoring of Insecticide Resistance Mutations in Native and Invasive Populations of the Fall Armyworm. Insects 2021, 12, 468. [Google Scholar] [CrossRef]
- Elias Oliveira Padovez, F.; Hideo Kanno, R.; Omoto, C.; Sartori Guidolin, A. Fitness Costs Associated with Chlorantraniliprole Resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) Strains with Different Genetic Backgrounds. Pest Manag. Sci. 2022, 78, 1279–1286. [Google Scholar] [CrossRef]
- Arias, O.; Cordeiro, E.; Corrêa, A.S.; Domingues, F.A.; Guidolin, A.S.; Omoto, C. Population Genetic Structure and Demographic History of Spodoptera frugiperda (Lepidoptera: Noctuidae): Implications for Insect Resistance Management Programs. Pest Manag. Sci. 2019, 75, 2948–2957. [Google Scholar] [CrossRef]


| Strain | N | LC50 (95% CI) a [μg a.i./cm2] | Slope ± SE | χ² | Df b | RR c | D |
|---|---|---|---|---|---|---|---|
| Sus | 768 | 0.05 (0.04–0.07) | 1.53 ± 0.11 | 16.2 | 9 | - | - |
| BA-R | 784 | >227 | 0.70 ± 0.26 | 2.8 | 10 | >4464 | - |
| Sus♀ × BA-R♂ | 832 | 0.11 (0.07–0.15) | 1.38 ± 0.12 | 18.1 | 9 | 2.12 | <−0.82 |
| Sus♂ × BA-R♀ | 832 | 0.15 (0.10–0.22) | 1.38 ± 0.13 | 26.3 | 10 | 2.98 | <−0.74 |
| TF-R | 512 | >227 | 0.80 ± 0.13 | 3.4 | 5 | >4464 | - |
| Sus♀ × TF-R♂ | 512 | 0.07 (0.05–0.10) | 1.53 ± 0.13 | 9.73 | 7 | 1.41 | <−0.92 |
| Sus♂ × TF-R♀ | 496 | 0.08 (0.05–0.11) | 1.29 ± 0.11 | 8.67 | 7 | 1.55 | <−0.89 |
| Flubendiamide (μg a.i./cm2) | Sus | BA-R ♂ × Sus ♀ | TF-R ♂ × Sus ♀ | BA-R | TF-R | ||
|---|---|---|---|---|---|---|---|
| Mortality (%) | Mortality (%) | Dominance (D) | Mortality (%) | Dominance (D) | Mortality (%) | Mortality (%) | |
| 0.00 | 2.08 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| 0.01 | 14.58 | 3.13 | 0.79 | 21.87 | 0.50 | 0.00 | 0.00 |
| 0.02 | 36.46 | 4.69 | 0.87 | 25.00 | 0.31 | 0.00 | 0.00 |
| 0.13 | 64.58 | 46.88 | 0.27 | 59.38 | 0.08 | 0.00 | 0.00 |
| 0.41 | 87.50 | 71.35 | 0.18 | 78.15 | 0.10 | 0.00 | 0.00 |
| 1.27 | 100.00 | 86.98 | 0.13 | 97.92 | 0.02 | 0.00 | 0.00 |
| 2.27 | 100.00 | 92.19 | 0.08 | 100.00 | 0.00 | 0.00 | 0.00 |
| 22.70 | 100.00 | 98.44 | 0.02 | 100.00 | 0.00 | 2.08 | 4.69 |
| 227.00 | 100.00 | 100.00 | 0.00 | 100.00 | 0.00 | 6.25 | 29.69 |
| Flubendiamide (μg a.i./cm2) | BA-R | TF-R | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BA-R ♀ x F1 ♂ | BA-R ♂ x F1 ♀ | TF-R ♀ x F1 ♂ | TF-R ♂ x F1 ♀ | |||||||||
| Obs. | Exp. | χ2 | Obs. | Exp. | χ2 | Obs. | Exp. | χ2 | Obs. | Exp. | χ2 | |
| 0.02 | 12.5 | 2.34 | 28.84 * | 10.42 | 2.34 | 18.22 * | 0 | 19.53 | 0.04 | 0 | 19.53 | 0.04 |
| 0.23 | 29.69 | 28.9 | 0.02 | 36.88 | 28.9 | 1.98 | 17.97 | 37.5 | 0.30 | 6.25 | 37.5 | 0.10 |
| 2.27 | 31.25 | 39.06 | 1.64 | 43.75 | 39.06 | 0.59 | 45.32 | 50 | 0.45 | 40.18 | 50 | 0.40 |
| 12.71 | 39.58 | 47.92 | 1.78 | 46.88 | 47.92 | 0.03 | 51.04 | 50 | 0.51 | 43.75 | 50 | 0.44 |
| 22.70 | 45.83 | 49.71 | 0.38 | 43.75 | 49.71 | 0.91 | 48.22 | 52.1 | 0.44 | 48.44 | 52.1 | 0.44 |
| 227.00 | 59.38 | 50.5 | 2.02 | 53.13 | 50.5 | 0.18 | 70 | 64.58 | 0.38 | 69.79 | 64.58 | 0.38 |
| Compound | Strain | n | LC50 (95% CI) a (µg a.i./cm²) | Slope ± SE | χ² | Df b | RR c |
|---|---|---|---|---|---|---|---|
| Chlorantraniliprole | Sus | 464 | 0.010 (0.007–0.014) | 1.33 ± 0.10 | 5.25 | 10 | - |
| BA-R | 496 | 7.13 (4.33–13.0) | 1.73 ± 0.08 | 18.2 | 8 | 713 | |
| TF-R | 448 | 40.4 (27.0–55.3) | 1.02 ± 0.08 | 10.6 | 7 | 4040 | |
| Cyantraniliprole | Sus | 224 | 0.002 (0.001–0.003) | 3.64 ± 0.66 | 0.74 | 6 | - |
| BA-R | 384 | 3.09 (0.59–6.29) | 1.62 ± 0.16 | 21.3 | 6 | 1545 | |
| TF-R | 800 | 12.8 (6.87–21.2) | 1.13 ± 0.07 | 12.8 | 7 | 6400 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okuma, D.M.; Cuenca, A.; Nauen, R.; Omoto, C. Large-Scale Monitoring of the Frequency of Ryanodine Receptor Target-Site Mutations Conferring Diamide Resistance in Brazilian Field Populations of Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2022, 13, 626. https://doi.org/10.3390/insects13070626
Okuma DM, Cuenca A, Nauen R, Omoto C. Large-Scale Monitoring of the Frequency of Ryanodine Receptor Target-Site Mutations Conferring Diamide Resistance in Brazilian Field Populations of Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects. 2022; 13(7):626. https://doi.org/10.3390/insects13070626
Chicago/Turabian StyleOkuma, Daniela M., Ana Cuenca, Ralf Nauen, and Celso Omoto. 2022. "Large-Scale Monitoring of the Frequency of Ryanodine Receptor Target-Site Mutations Conferring Diamide Resistance in Brazilian Field Populations of Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae)" Insects 13, no. 7: 626. https://doi.org/10.3390/insects13070626







