The Genomics of Isolated Populations of Gampsocleis glabra (Orthoptera: Tettigoniidae) in Central and Western Europe
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
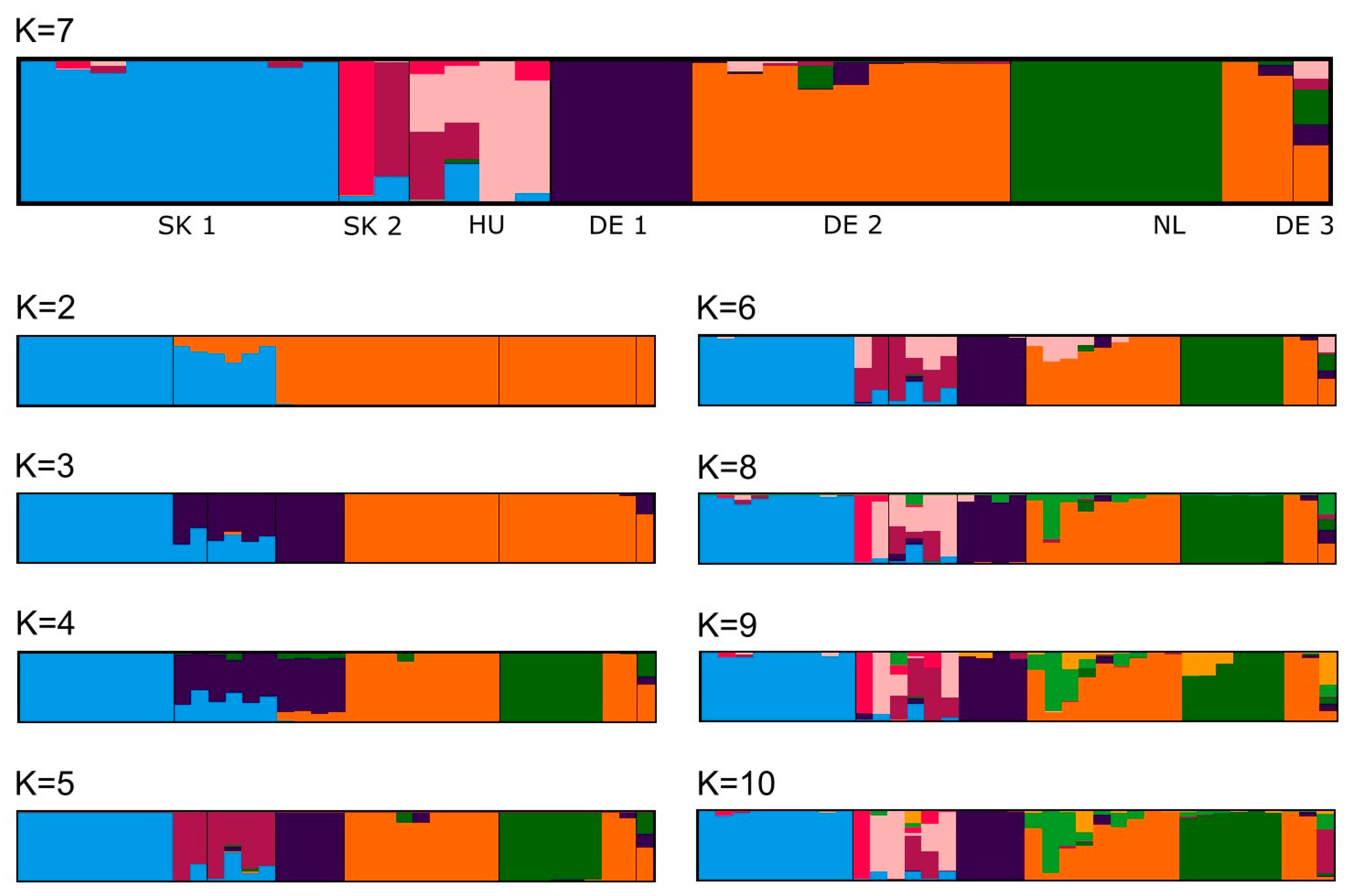
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Locality | Code | Latitude | Longitude | Description |
|---|---|---|---|---|
| Veľký Kamenec | SK 1 | 48.3741 | 21.8254 | Sand dunes with high grass |
| Strážne | SK 2 | 48.3875 | 21.8465 | Cow pasture with high grass |
| Kócsujfalu | HU | 47.5703 | 20.9444 | Puszta meadows, pasture |
| Klietz | DE 1 | 52.6451 | 12.1318 | Calluna heathland, military training |
| Rheinmetall | DE 2 | 52.8829 | 10.2861 | Calluna heathland, military training |
| Oldebroek | NL | 52.4194 | 5.9575 | Calluna heathland, military training |
| Munster | DE 3 | 52.9451 | 10.0502 | Calluna heathland, military training |
References
- Pereira, H.M.; Leadley, P.W.; Proença, V.; Alkemade, R.; Scharlemann, J.P.W.; Fernandez-Manjarrés, J.F.; Araújo, M.B.; Balvanera, P.; Biggs, R.; Cheung, W.W.L.; et al. Scenarios for Global Biodiversity in the 21st Century. Science (1979) 2010, 330, 1496–1501. [Google Scholar] [CrossRef]
- Pardini, R.; Nichols, E.; Püttker, T. Biodiversity Response to Habitat Loss and Fragmentation. Encycl. Anthr. 2017, 3, 229–239. [Google Scholar] [CrossRef]
- Hoekstra, J.M.; Boucher, T.M.; Ricketts, T.H.; Roberts, C. Confronting a Biome Crisis: Global Disparities of Habitat Loss and Protection. Ecol. Lett. 2005, 8, 23–29. [Google Scholar] [CrossRef]
- Wilson, M.C.; Chen, X.Y.; Corlett, R.T.; Didham, R.K.; Ding, P.; Holt, R.D.; Holyoak, M.; Hu, G.; Hughes, A.C.; Jiang, L.; et al. Habitat Fragmentation and Biodiversity Conservation: Key Findings and Future Challenges. Landsc. Ecol. 2016, 31, 219–227. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of Habitat Fragmentation on Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Louy, D.; Habel, J.C.; Schmitt, T.; Assmann, T.; Meyer, M.; Müller, P. Strongly Diverging Population Genetic Patterns of Three Skipper Species: The Role of Habitat Fragmentation and Dispersal Ability. Conserv. Genet. 2007, 8, 671–681. [Google Scholar] [CrossRef]
- Dey, L.S.; Simões, M.V.P.; Hawlitschek, O.; Sergeev, M.G.; Xu, S.Q.; Lkhagvasuren, D.; Husemann, M. Analysis of Geographic Centrality and Genetic Diversity in the Declining Grasshopper Species Bryodemella tuberculata (Orthoptera: Oedipodinae). Biodivers. Conserv. 2021, 30, 2773–2796. [Google Scholar] [CrossRef]
- Gibson, D.J. Grasses and Grassland Ecology; Oxford University Press: Oxford, UK, 2009; ISBN 9780191546099. [Google Scholar]
- Török, P.; Neuffer, B.; Heilmeier, H.; Bernhardt, K.G.; Wesche, K. Climate, Landscape History and Management Drive Eurasian Steppe Biodiversity. Flora 2020, 271, 151685. [Google Scholar] [CrossRef]
- Kajtoch, Ł.; Cieślak, E.; Varga, Z.; Paul, W.; Mazur, M.A.; Sramkó, G.; Kubisz, D. Phylogeographic Patterns of Steppe Species in Eastern Central Europe: A Review and the Implications for Conservation. Biodivers. Conserv. 2016, 25, 2309–2339. [Google Scholar] [CrossRef]
- Reif, J.; Chajma, P.; Dvořáková, L.; Koptík, J.; Marhoul, P.; Čížek, O.; Kadlec, T. Biodiversity Changes in Abandoned Military Training Areas: Relationships to Different Management Approaches in Multiple Taxa. Front. Environ. Sci. 2023, 01-18. [Google Scholar] [CrossRef]
- Porter, E.E.; Redak, R.A.; Braker, H.E. Density, Biomass, and Diversity of Grasshoppers (Orthoptera: Acrididae) in a California Native Grassland. Great Basin Nat. 1996, 56, 172–176. [Google Scholar]
- Branson, D.H.; Joern, A.; Sword, G.A. Sustainable Management of Insect Herbivores in Grassland Ecosystems: New Perspectives in Grasshopper Control. Bioscience 2006, 56, 743–755. [Google Scholar] [CrossRef]
- Cigliano, M.M.; Braun, H.; Eades, D.C.; Otte, D. Orthoptera Species File Version 5.0/5.0. Available online: https://orthoptera.speciesfile.org/ (accessed on 1 November 2023).
- Herbst, J.W. Forsetzung Des Verzeichnisses Meiner Insektensammlung. In Archiv Der Insectengeschichte; Fuessly, J.C., Ed.; Fuessli: Zürich, Switzerland, 1786; pp. 183–196. [Google Scholar]
- Bellmann, H.; Rutschmann, F.; Roesti, C.; Hochkirch, A. Der Kosmos Heuschrecken-Führer; Kosmos: Stuttgart, Germany, 2019. [Google Scholar]
- Hochkirch, A. Gampsocleis glabra (Steppe Spiny Bush-Cricket). Available online: https://www.iucnredlist.org/species/44711951/115472224 (accessed on 12 September 2023).
- Schäfer, B. Nachweis Der Heideschrecke Gampsocleis glabra (Herbst, 1786) (Ensifera) in Der Colbitz-Letzlinger Heide (Sachsen-Anhalt). Articulata 2013, 28, 115–126. [Google Scholar]
- Schäfer, B.; Hennigs, S. Nachweise Der Heideschrecke Gampsocleis glabra (Herbst, 1786) (Ensifera) in Der Altengrabower Sowie in Der Klietzer Heide (Brandenburg/Sachsen-Anhalt). Articulata 2020, 35, 117–127. [Google Scholar]
- van der Berg, A.; Haveman, R.; Hornman, M. De Kleine Wrattenbijter Gampsocleis glabra Herontdekt in Nederland (Orthoptera: Tettigoniidae). Ned. Faun. Meded. 2000, 2, 1–12. [Google Scholar]
- Sardet, E.; Roesti, C.; Braud, Y. Cahier d’identification Des Orthoptères de France, Belgique, Luxembourg & Suisse; Biotope: Mèze, France, 2015; ISBN 9782366621556. [Google Scholar]
- Grzędzicka, E.; Vahed, K. Habitat Requirements of the Endangered Heath Bush-Cricket Gampsocleis glabra (Orthoptera, Tettigoniidae) in an Isolated Population. J. Insect Conserv. 2020, 24, 935–945. [Google Scholar] [CrossRef]
- Fedor, P.; Holuša, J.; Majzlan, O.; Prokop, P. Distribution, Conservation and Prognosis for Gampsocleis glabra (Herbst, 1786) (Insecta: Ensifera) in Slovakia and the Czech Republic. Articulata 2004, 19, 217–224. [Google Scholar]
- Krištín, A.; Kaňuch, P.; Balla, M.; Gavlas, V. On the Distribution and Ecology of Gampsocleis glabra and Tettigonia caudata (Orthoptera) in Slovakia. Articulata 2007, 22, 53–61. [Google Scholar]
- Vogels, J.J.; Van de Waal, D.B.; Wallis de Vries, M.F.; Van den Burg, A.B.; Nijssen, M.; Bobbink, R.; Berg, M.P.; Olde Venterink, H.; Siepel, H. Towards a Mechanistic Understanding of the Impacts of Nitrogen Deposition on Producer–Consumer Interactions. Biol. Rev. 2023, 98, 1712–1731. [Google Scholar] [CrossRef]
- Vogels, J.J.; Verberk, W.C.E.P.; Lamers, L.P.M.; Siepel, H. Can Changes in Soil Biochemistry and Plant Stoichiometry Explain Loss of Animal Diversity of Heathlands? Biol. Conserv. 2017, 212, 432–447. [Google Scholar] [CrossRef]
- Roesti, C.; Rutschmann, F. Orthoptera.Ch. Available online: http://www.orthoptera.ch/ (accessed on 12 September 2023).
- Pierson, J.C.; Beissinger, S.R.; Bragg, J.G.; Coates, D.J.; Gerard, J.; Oostermeijer, B.; Sunnucks, P.; Schumaker, N.H.; Trotter, M.V.; Young, A.G. Incorporating Evolutionary Processes into Population Viability Models. Conserv. Biol. 2015, 29, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Formenti, G.; Theissinger, K.; Fernandes, C.; Bista, I.; Bombarely, A.; Bleidorn, C.; Ciofi, C.; Crottini, A.; Godoy, J.A.; Höglund, J.; et al. The Era of Reference Genomes in Conservation Genomics. Trends Ecol. Evol. 2022, 37, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Hawlitschek, O.; Sadílek, D.; Dey, L.S.; Buchholz, K.; Noori, S.; Baez, I.L.; Wehrt, T.; Brozio, J.; Trávníček, P.; Seidel, M.; et al. New Estimates of Genome Size in Orthoptera and Their Evolutionary Implications. PLoS ONE 2023, 18, e0275551. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for de Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Escoda, L.; Hawlitschek, O.; González-Esteban, J.; Castresana, J. Methodological Challenges in the Genomic Analysis of an Endangered Mammal Population with Low Genetic Diversity. Sci. Rep. 2022, 12, 21390. [Google Scholar] [CrossRef]
- Çilingir, F.G.; Hansen, D.; Ozgul, A.; Grossen, C. Design of SNP Markers for Aldabra Giant Tortoises Using Low Coverage DdRAD-Seq. Conserv. Genet. Resour. 2021, 13, 409–412. [Google Scholar] [CrossRef]
- Roy, S.C.; Moitra, K.; De Sarker, D. Assessment of Genetic Diversity among Four Orchids Based on DdRAD Sequencing Data for Conservation Purposes. Physiol. Mol. Biol. Plants 2017, 23, 169–183. [Google Scholar] [CrossRef]
- Querejeta, M.; González-Esteban, J.; Gómez, A.; Fernández-González, A.; Aymerich, P.; Gosálbez, J.; Escoda, L.; Igea, J.; Castresana, J. Genomic Diversity and Geographical Structure of the Pyrenean Desman. Conserv. Genet. 2016, 17, 1333–1344. [Google Scholar] [CrossRef]
- Noguerales, V.; Ortego, J. Genomic Evidence of Speciation by Fusion in a Recent Radiation of Grasshoppers. Evolution 2022, 76, 2618–2633. [Google Scholar] [CrossRef]
- González-Serna, M.J.; Cordero, P.J.; Ortego, J. Using High-Throughput Sequencing to Investigate the Factors Structuring Genomic Variation of a Mediterranean Grasshopper of Great Conservation Concern. Sci. Rep. 2018, 8, 13436. [Google Scholar] [CrossRef]
- Paxton, R.J.; Thorén, P.A.; Tengö, J.; Estoup, A.; Pamilo, P. Mating Structure and Nestmate Relatedness in a Communal Bee, Andrena Jacobi (Hymenoptera, Andrenidae), Using Microsatellites. Mol. Ecol. 1996, 5, 511–519. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An Analysis Tool Set for Population Genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and Genotyping Loci de Novo from Short-Read Sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New Tools for the Analysis of Genome-Wide SNP Data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. Structure Harvester: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Hagberg, L.; Celemín, E.; Irisarri, I.; Hawlitschek, O.; Bella, J.L.; Mott, T.; Pereira, R.J.; Ricardo Pereira, C.J. Extensive Introgression at Late Stages of Species Formation: Insights from Grasshopper Hybrid Zones. Mol. Ecol. 2022, 31, 2384–2399. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, E.M.; Sari, M.; Başibüyük, H.H. Genetic Structure of Chorthippus parallelus (Orthoptera: Acrididae: Gomphocerinae) Populations in Anatolia: A Stable Rear Edge Population. Ann. Entomol. Soc. Am. 2010, 103, 625–634. [Google Scholar] [CrossRef]
- Pecsenye, K.; Vadkerti, E.; Academiae, Z.V.-A.Z. Temporal and Spatial Pattern of Genetic Differentiation in Isophya kraussii (Orthoptera: Tettigonoidea) in NE Hungary. Acta Zool. Acad. Sci. Hung. 2003, 49, 167–178. [Google Scholar]
- Pecsenye, K.; Vadkerti, E.; Varga, Z. Pattern of Genetic Differentiation in Two Isophya Species (Orthoptera: Tettigonoidea) in North-East Hungary. J. Insect Conserv. 2003, 7, 207–213. [Google Scholar] [CrossRef]
- Xu, Y.; Mai, J.; Yu, B.; Hu, H.; Yuan, L.; Jashenko, R.; Ji, R. Study on the Genetic Differentiation of Geographic Populations of Calliptamus italicus (Orthoptera: Acrididae) in Sino-Kazakh Border Areas Based on Mitochondrial COI. J. Econ. Entomol. 2019, 112, 1912–1919. [Google Scholar] [CrossRef]
- Nolen, Z.J.; Yildirim, B.; Irisarri, I.; Liu, S.; Groot Crego, C.; Amby, D.B.; Mayer, F.; Gilbert, M.T.P.; Pereira, R.J. Historical Isolation Facilitates Species Radiation by Sexual Selection: Insights from Chorthippus Grasshoppers. Mol. Ecol. 2020, 29, 4985–5002. [Google Scholar] [CrossRef]
- Kelemen, A.; Török, P.; Valkó, O.; Deák, B.; Miglécz, T.; Tóth, K.; Ölvedi, T.; Tóthmérész, B. Sustaining Recovered Grasslands Is Not Likely without Proper Management: Vegetation Changes after Cessation of Mowing. Biodivers. Conserv. 2014, 23, 741–751. [Google Scholar] [CrossRef]
- Harz, K. Die Geradflügler Mitteleuropas; Fischer, G., Ed.; Fischer: Jena, Germany, 1957. [Google Scholar]
- Schlumprecht, H.; Waeber, G. Heuschrecken in Bayern; Ulmer: Stuttgart, Germany, 2003. [Google Scholar]

| No. | Sex | Collection Date | Collector | Country | Locality |
|---|---|---|---|---|---|
| ML20 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML21 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML23 | m | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML26 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML27 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML29 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML30 | m | 13 August 2020 | OH | Slovakia | Strážne (SK 2) |
| ML32 | m | 13 August 2020 | OH | Hungary | Kócsujfalu (HU) |
| ML33 | m | 13 August 2020 | OH | Hungary | Kócsujfalu (HU) |
| ML34 | f | 13 August 2020 | OH | Hungary | Kócsujfalu (HU) |
| ML35 | m | 13 August 2020 | OH | Hungary | Kócsujfalu (HU) |
| ML52 | m | 17 August 2020 | MH | Germany | Klietz (DE 1) |
| ML54 | m | 17 August 2020 | MH | Germany | Klietz (DE 1) |
| ML58 | m | 17 August 2020 | MH | Germany | Klietz (DE 1) |
| ML62 | m | 17 August 2020 | MH | Germany | Klietz (DE 1) |
| ML70 | f | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML71 | f | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML73 | f | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML75 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML76 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML77 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML88 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML89 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML90 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML92 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML93 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML94 | m | 31 July 2020 | RF, HvK | The Netherlands | Oldebroek (NL) |
| ML99 | m | 13 August 2020 | OH | Slovakia | Strážne (SK 2) |
| ML100 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML101 | m | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML102 | f | 13 August 2020 | OH | Slovakia | Veľký Kamenec (SK 1) |
| ML104 | m | 27 August 2020 | MH | Germany | Munster (DE 3) |
| ML109 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML110 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML112 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML114 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| ML115 | m | 28 August 2020 | MH | Germany | Rheinmetall (DE 2) |
| Pop. | SK 1 | SK 2 | HU | DE 1 | DE 2 | NL | DE 3 |
|---|---|---|---|---|---|---|---|
| N | 9 | 2 | 4 | 4 | 11 | 6 | 1 |
| SK 2 | 0.036 | ||||||
| HU | 0.139 | 0.106 | |||||
| DE 1 | 0.124 | 0.086 | 0.090 | ||||
| DE 2 | 0.115 | 0.081 | 0.081 | 0.037 | |||
| NL | 0.010 | 0.031 | 0.139 | 0.121 | 0.108 | ||
| DE 3 | 0.165 | 0.068 | 0.147 | 0.038 | 0.051 | 0.181 | |
| FIS | 0.523 | 0.235 | 0.184 | 0.600 | 0.463 | 0.282 | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawlitschek, O.; Bruns, C.; Dey, L.-S.; Nuhlíčková, S.; Felix, R.; van Kleef, H.; Nakel, J.; Husemann, M. The Genomics of Isolated Populations of Gampsocleis glabra (Orthoptera: Tettigoniidae) in Central and Western Europe. Insects 2023, 14, 946. https://doi.org/10.3390/insects14120946
Hawlitschek O, Bruns C, Dey L-S, Nuhlíčková S, Felix R, van Kleef H, Nakel J, Husemann M. The Genomics of Isolated Populations of Gampsocleis glabra (Orthoptera: Tettigoniidae) in Central and Western Europe. Insects. 2023; 14(12):946. https://doi.org/10.3390/insects14120946
Chicago/Turabian StyleHawlitschek, Oliver, Carsten Bruns, Lara-Sophie Dey, Soňa Nuhlíčková, Rob Felix, Hein van Kleef, Jacqueline Nakel, and Martin Husemann. 2023. "The Genomics of Isolated Populations of Gampsocleis glabra (Orthoptera: Tettigoniidae) in Central and Western Europe" Insects 14, no. 12: 946. https://doi.org/10.3390/insects14120946





