Dehydration Alters Transcript Levels in the Mosquito Midgut, Likely Facilitating Rapid Rehydration following a Bloodmeal
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hagan, R.W.; Didion, E.M.; Rosselot, A.E.; Holmes, C.J.; Siler, S.C.; Rosendale, A.J.; Hendershot, J.M.; Elliot, K.S.B.; Jennings, E.C.; Nine, G.A.; et al. Dehydration Prompts Increased Activity and Blood Feeding by Mosquitoes. Sci. Rep. 2018, 8, 6804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canyon, D.V.; Hii, J.L.K.; Müller, R. Adaptation of Aedes aegypti (Diptera: Culicidae) Oviposition Behavior in Response to Humidity and Diet. J. Insect Physiol. 1999, 45, 959–964. [Google Scholar] [CrossRef]
- Costa, E.A.P.D.A.; Santos, E.M.D.M.; Correia, J.C.; Albuquerque, C.M.R.D. Impact of Small Variations in Temperature and Humidity on the Reproductive Activity and Survival of Aedes aegypti (Diptera, Culicidae). Rev. Bras. Entomol. 2010, 54, 488–493. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.A.; Maibach, H.I. A Study of the Probing Response of Aedes aegypti. 4. Effect of Dry and Moist Heat on Probing. J. Econ. Entomol. 1971, 64, 442–443. [Google Scholar] [CrossRef] [Green Version]
- Rowley, W.A.; Graham, C.L. The Effect of Temperature and Relative Humidity on the Flight Performance of Female Aedes aegypti. J. Insect Physiol. 1968, 14, 1251–1257. [Google Scholar] [CrossRef]
- Parker, A.H. The Effect of a Difference in Temperature and Humidity on Certain Reactions of Female Aedes aegypti (L.). Bull. Entomol. Res. 1952, 43, 221–229. [Google Scholar] [CrossRef]
- Holmes, C.J.; Brown, E.S.; Sharma, D.; Nguyen, Q.; Spangler, A.A.; Pathak, A.; Payton, B.; Warden, M.; Shah, A.J.; Shaw, S.; et al. Bloodmeal Regulation in Mosquitoes Curtails Dehydration-Induced Mortality, Altering Vectorial Capacity. J. Insect Physiol. 2022, 137, 104363. [Google Scholar] [CrossRef]
- Benoit, J.B.; Patrick, K.R.; Desai, K.; Hardesty, J.J.; Krause, T.B.; Denlinger, D.L. Repeated Bouts of Dehydration Deplete Nutrient Reserves and Reduce Egg Production in the Mosquito Culex pipiens. J. Exp. Biol. 2010, 213, 2763–2769. [Google Scholar] [CrossRef] [Green Version]
- Reidenbach, K.R.; Cheng, C.; Liu, F.; Liu, C.; Besansky, N.J.; Syed, Z. Cuticular Differences Associated with Aridity Acclimation in African Malaria Vectors Carrying Alternative Arrangements of Inversion 2La. Parasites Vectors 2014, 7, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canyon, D.V.; Muller, R.; Hii, J.L.K. Aedes aegypti Disregard Humidity-Related Conditions with Adequate Nutrition. Trop. Biomed. 2013, 30, 1–8. [Google Scholar]
- Dow, R.P.; Gerrish, G.M. Day-to-Day Change in Relative Humidity and the Activity of Culex nigripalpus (Diptera: Culicidae). Ann. Entomol. Soc. Am. 1970, 63, 995–999. [Google Scholar] [CrossRef]
- Lyons, C.L.; Coetzee, M.; Terblanche, J.S.; Chown, S.L. Desiccation Tolerance as a Function of Age, Sex, Humidity and Temperature in Adults of the African Malaria Vectors Anopheles arabiensis and Anopheles funestus. J. Exp. Biol. 2014, 217, 3823–3833. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M. Effect of Temperature and Humidity on Life Cycle Duration of Culex quinquefasciatus Say (Diptera : Culicidae) at Muzaffarpur (Bihar), India. Adv. Biores. 2015, 6, 103–105. [Google Scholar] [CrossRef]
- Leeson, H.S. Longevity of Anopheles maculipennis Race Atroparvus, van Thiel, at Controlled Temperature and Humidity after One Blood Meal. Bull. Entomol. Res. 1939, 30, 295–301. [Google Scholar] [CrossRef]
- Mayne, B. Notes on the Influence of Temperature and Humidity on Oviposition and Early Life of Anopheles. Public Health Rep. 1926, 41, 986–990. [Google Scholar] [CrossRef]
- Ruiz, M.O.; Chaves, L.F.; Hamer, G.L.; Sun, T.; Brown, W.M.; Walker, E.D.; Haramis, L.; Goldberg, T.L.; Kitron, U.D. Local Impact of Temperature and Precipitation on West Nile Virus Infection in Culex Species Mosquitoes in Northeast Illinois, USA. Parasites Vectors 2010, 3, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Holmes, C.J.; Benoit, J.B. Biological Adaptations Associated with Dehydration in Mosquitoes. Insects 2019, 10, 375. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, R.; Fleisher, A.E.; Minnick, S.L.; Simmons, K.A.; Scott, T.W. Nutritional Stress Affects Mosquito Survival and Vector Competence for West Nile Virus. Vector-Borne Zoonotic Dis. 2008, 8, 727–732. [Google Scholar] [CrossRef]
- Briegel, H.; Hörler, E. Multiple Blood Meals as a Reproductive Strategy in Anopheles (Diptera: Culicidae). J. Med. Entomol. 1993, 30, 975–985. [Google Scholar] [CrossRef] [Green Version]
- Lea, A.O.; Briegel, H.; Lea, H.M. Arrest, Resorption, or Maturation of Oocytes in Aedes aegypti: Dependence on the Quantity of Blood and the Interval between Blood Meals. Physiol. Entomol. 1978, 3, 309–316. [Google Scholar] [CrossRef]
- Paz, S. Climate Change Impacts on West Nile Virus Transmission in a Global Context. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20130561. [Google Scholar] [CrossRef]
- Dodson, B.L.; Rasgon, J.L. Vector Competence of Anopheles and Culex Mosquitoes for Zika Virus. PeerJ 2017, 5, e3096. [Google Scholar] [CrossRef] [Green Version]
- Tingström, O.; Wesula Lwande, O.; Näslund, J.; Spyckerelle, I.; Engdahl, C.; Von Schoenberg, P.; Ahlm, C.; Evander, M.; Bucht, G. Detection of Sindbis and Inkoo Virus RNA in Genetically Typed Mosquito Larvae Sampled in Northern Sweden. Vector-Borne Zoonotic Dis. 2016, 16, 461–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Dong, Y.; Huang, Y.; Rasgon, J.L.; Agre, P. Impact of Trehalose Transporter Knockdown on Anopheles gambiae Stress Adaptation and Susceptibility to Plasmodium falciparum Infection. Proc. Natl. Acad. Sci. USA 2013, 110, 17504–17509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naksathit, A.T.; Edman, J.D.; Scott, T.W. Utilization of Human Blood and Sugar as Nutrients by Female Aedes aegypti (Diptera: Culicidae). J. Med. Entomol 1999, 36, 13–17. [Google Scholar] [CrossRef]
- Drake, L.L.; Boudko, D.Y.; Marinotti, O.; Carpenter, V.K.; Dawe, A.L.; Hansen, I.A. The Aquaporin Gene Family of the Yellow Fever Mosquito, Aedes aegypti. PLoS ONE 2010, 5, e15578. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.C.; Hagedorn, H.H.; Beyenbach, K.W. Dynamic Changes in Flow Rate and Composition of Urine during the Post-Bloodmeal Diuresis in Aedes aegypti (L.). J. Comp. Physiol. B 1983, 153, 257–265. [Google Scholar] [CrossRef]
- Matthews, B.J.; Dudchenko, O.; Kingan, S.B.; Koren, S.; Antoshechkin, I.; Crawford, J.E.; Glassford, W.J.; Herre, M.; Redmond, S.N.; Rose, N.H.; et al. Improved Reference Genome of Aedes aegypti Informs Arbovirus Vector Control. Nature 2018, 563, 501–507. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput. 2015. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 February 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Mount, S.M.; Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms Nat. Biotechnol. 2014, 32, 462–464. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. G:Profiler-a Web-Based Toolset for Functional Profiling of Gene Lists from Large-Scale Experiments. Nucleic Acids Res. 2007, 35, 193–200. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsova, I.; Lugmayr, A.; Siira, S.J.; Rackham, O.; Filipovska, A. CirGO: An Alternative Circular Way of Visualising Gene Ontology Terms. BMC Bioinform. 2019, 20, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting Differential Usage of Exons from RNA-Seq Data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Detinova, T.S.; Beklemishev, W.N.; Bertram, D.S. Age-Grouping Methods in Diptera of Medical Importance With Special Reference to Some Vectors of Malaria. World Health Organ. Monogr. Ser. 1962, 47, 1–213. [Google Scholar] [CrossRef]
- The GIMP Development Team GIMP. Available online: https://www.gimp.org (accessed on 26 February 2023).
- Rivers, D.B.; Denlinger, D.L. Redirection of Metabolism in the Flesh Fly, Sarcophaga bullata, Following Envenomation by the Ectoparasitoid Nasonia vitripennis and Correlation of Metabolic Effects with the Diapause Status of the Host. J. Insect Physiol. 1994, 40, 207–215. [Google Scholar] [CrossRef]
- Van Handel, E. Rapid Determination of Glycogen and Sugars in Mosquitoes. J. Am. Mosq. Control Assoc. 1985, 1, 299–301. [Google Scholar] [PubMed]
- Van Handel, E. Rapid Determination of Total Lipids in Mosquitoes. J. Am. Mosq. Control Assoc. 1985, 1, 302–304. [Google Scholar] [PubMed]
- Microsoft Corporation Microsoft Excel. Available online: https://office.microsoft.com/excel (accessed on 26 February 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Tidyr: Tidy Messy Data 2020. Available online: https://tidyr.tidyverse.org (accessed on 26 February 2023).
- Wickham, H.; Francois, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation 2017. Available online: https://dplyr.tidyverse.org (accessed on 26 February 2023).
- Hope, R.M. Rmisc: Rmisc: Ryan Miscellaneous 2013. Available online: https://cran.r-project.org/package=Rmisc (accessed on 26 February 2023).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Inkscape Project. Inkscape 2020. Available online: https://inkscape.org (accessed on 26 February 2023).
- Li, Y.; Piermarini, P.M.; Esquivel, C.J.; Drumm, H.E.; Schilkey, F.D.; Hansen, I.A. RNA-Seq Comparison of Larval and Adult Malpighian Tubules of the Yellow Fever Mosquito Aedes aegypti Reveals Life Stage-Specific Changes in Renal Function. Front. Physiol. 2017, 8, 283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayar, J.K.; Sauerman, D.M. The Effects of Nutrition on Survival and Fecundity on Florida Mosquitoes. J. Med. Ent 1975, 12, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, M.C.; Wides, R.; et al. The Genome Sequence of the Malaria Mosquito Anopheles gambiae. October 2002, 298, 129–149. [Google Scholar] [CrossRef]
- Foster, W.A. Mosquito Sugar Feeding and Reproductive Energetics. Annu. Rev. Entomol. 1995, 40, 443–474. [Google Scholar] [CrossRef]
- Lehane, M.J. The Biology of Blood-Sucking in Insects, Second Edition; Cambridge University Press: New York, NY, USA, 2005; ISBN 9780511610493. [Google Scholar]
- Sanders, H.R.; Foy, B.D.; Evans, A.M.; Ross, L.S.; Beaty, B.J.; Olson, K.E.; Gill, S.S. Sindbis Virus Induces Transport Processes and Alters Expression of Innate Immunity Pathway Genes in the Midgut of the Disease Vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2005, 35, 1293–1307. [Google Scholar] [CrossRef]
- Drake, L.L.; Rodriguez, S.D.; Hansen, I.A. Functional Characterization of Aquaporins and Aquaglyceroporins of the Yellow Fever Mosquito, Aedes aegypti. Sci. Rep. 2015, 5, 7795. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Tsujimoto, H.; Cha, S.S.-J.; Agre, P.; Rasgon, J.L. Aquaporin Water Channel AgAQP1 in the Malaria Vector Mosquito Anopheles gambiae during Blood Feeding and Humidity Adaptation. Proc. Natl. Acad. Sci. USA 2011, 108, 6062–6066. [Google Scholar] [CrossRef] [Green Version]
- Tsujimoto, H.; Liu, K.; Linser, P.J.; Agre, P.; Rasgon, J.L. Organ-Specific Splice Variants of Aquaporin Water Channel AgAQP1 in the Malaria Vector Anopheles gambiae. PLoS ONE 2013, 8, e75888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piermarini, P.M. Renal Excretory Processes in Mosquitoes, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2016; Volume 51, ISBN 9780128024577. [Google Scholar]
- Van Handel, E.; Romoser, W.S. Proteolytic Activity in the Ectoperitrophic Fluid of Blood-fed Culex nigripalpus. Med. Vet. Entomol. 1987, 1, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.N. The Biology of Mosquitoes: Development, Nutrition and Reproduction; Chapman & Hall: London, UK, 1992; Volume 1, ISBN 9780412401800. [Google Scholar]
- Brugier, A.; Hafirrassou, M.L.; Pourcelot, M.; Baldaccini, M.; Kril, V.; Couture, L.; Kümmerer, B.M.; Gallois-Montbrun, S.; Bonnet-Madin, L.; Vidalain, P.-O.; et al. RACK1 Associates with RNA-Binding Proteins Vigilin and SERBP1 to Facilitate Dengue Virus Replication. J. Virol. 2022, 96, e01962-21. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, P.M.; Ehrlich, H.Y.; Magalhaes, T.; Miller, M.R.; Conway, P.J.; Bransfield, A.; Misencik, M.J.; Gloria-Soria, A.; Warren, J.L.; Andreadis, T.G.; et al. Successive Blood Meals Enhance Virus Dissemination within Mosquitoes and Increase Transmission Potential. Nat. Microbiol. 2020, 5, 239–247. [Google Scholar] [CrossRef]

| Group | Dehydration | Bloodfed | Sample | Paired-End Reads |
|---|---|---|---|---|
| N1 | No | No | N1-2 | 75,496,800 |
| N1-3 | 88,761,156 | |||
| Y1 | No | Yes | Y1-1 | 89,322,470 |
| Y1-2 | 81,691,584 | |||
| Y1-3 | 74,120,060 | |||
| N7 | Yes | No | N7-1 | 105,818,594 |
| N7-2 | 105,385,942 | |||
| N7-3 | 82,647,520 | |||
| Y7 | Yes | Yes | Y7-1 | 95,531,032 |
| Y7-2 | 85,314,086 |
| Group | Comparison | Genes | GO Pathways | REVIGO Terms |
|---|---|---|---|---|
| Y1N1 | Y1/N1 | 145 | 7 | 4 |
| N1/Y1 | 62 | 3 | 3 | |
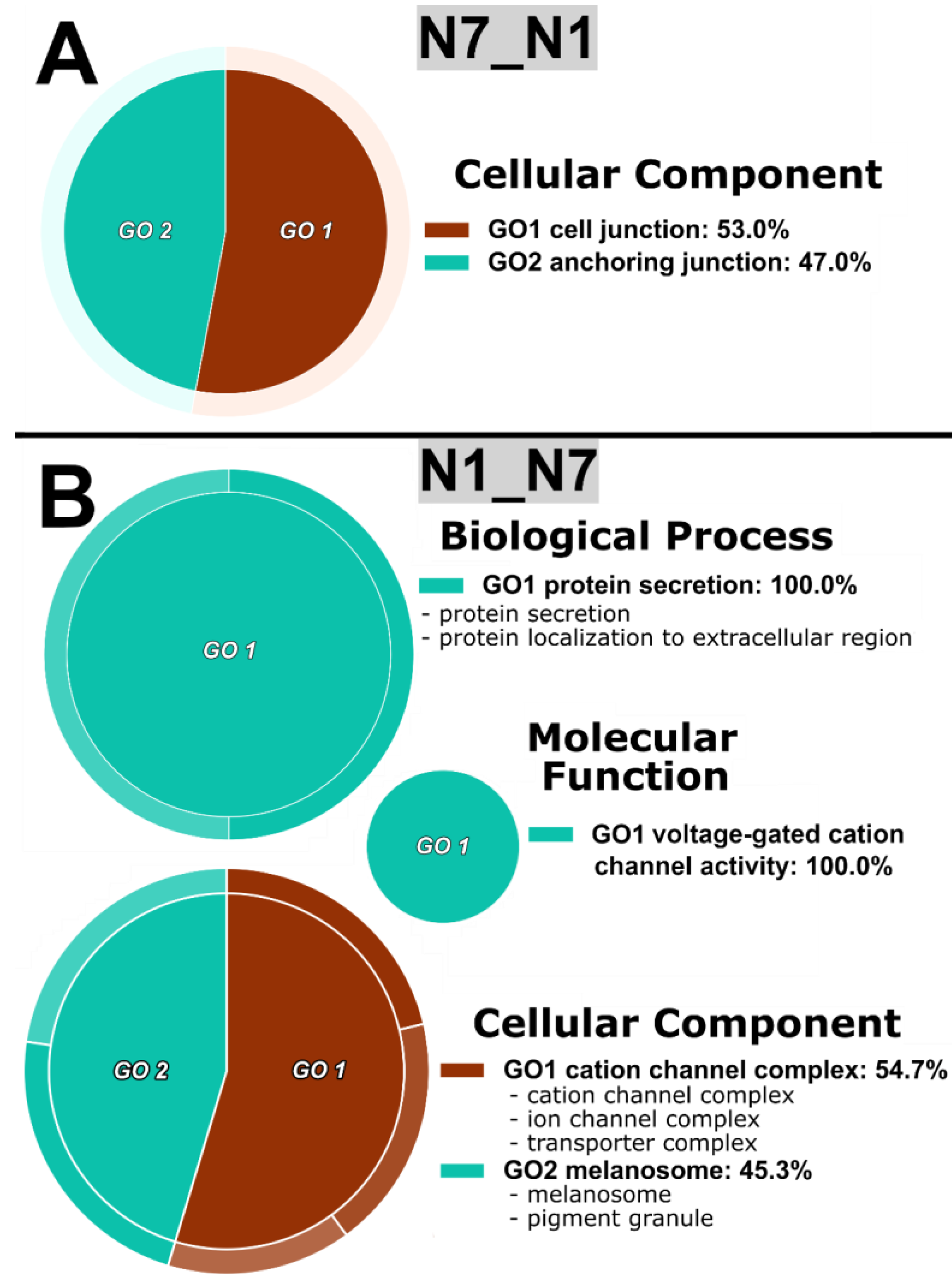
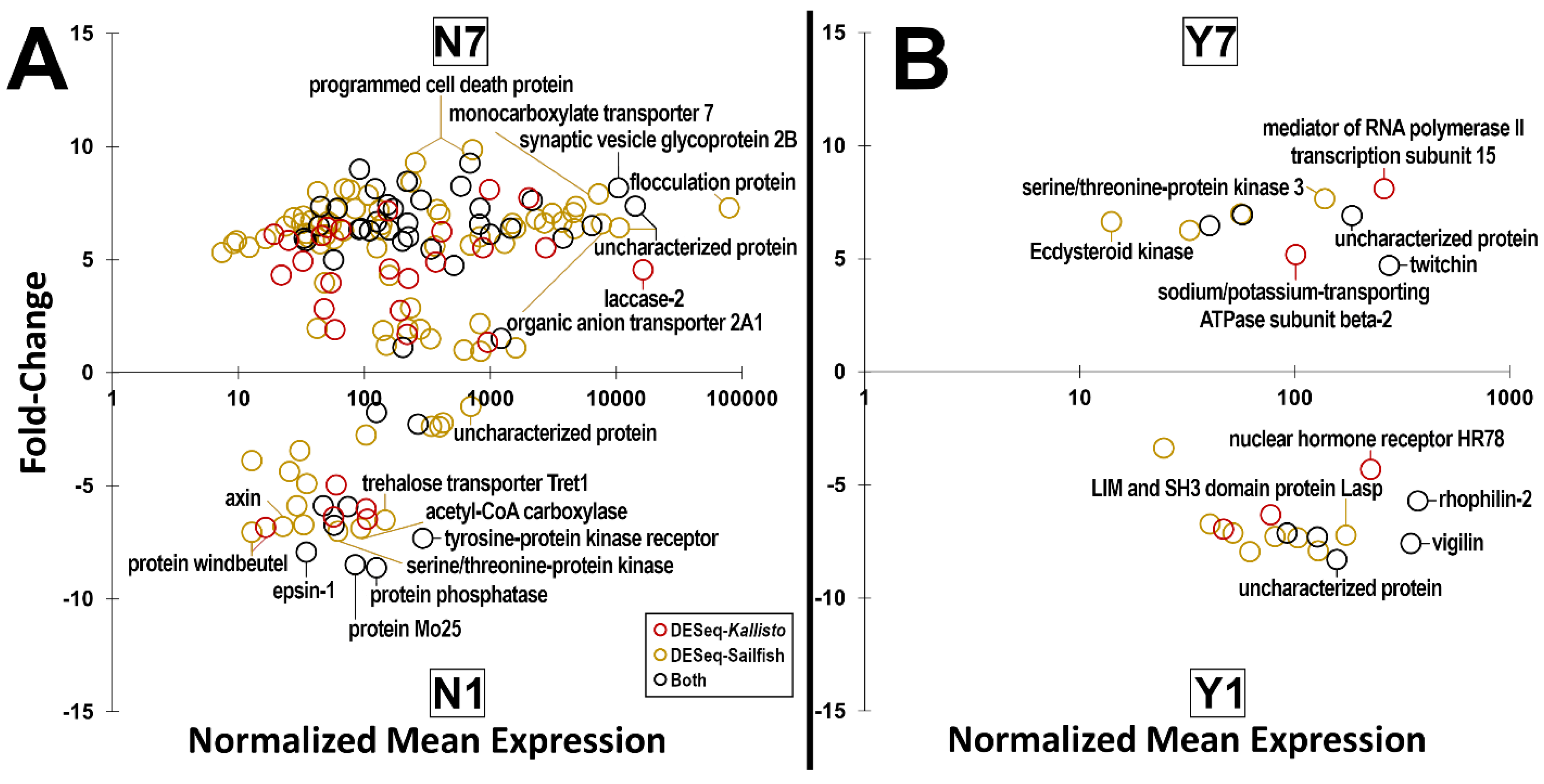
| N7N1 | N7/N1 | 146 | 4 | 2 |
| N1/N7 | 91 | 13 | 5 | |
| Y7Y1 | Y7/Y1 | 37 | 0 | 0 |
| Y1/Y7 | 40 | 0 | 0 | |
| N7Y7 | N7/Y7 | 390 | 8 | 5 |
| Y7/N7 | 281 | 29 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holmes, C.J.; Brown, E.S.; Sharma, D.; Warden, M.; Pathak, A.; Payton, B.; Nguyen, Q.; Spangler, A.; Sivakumar, J.; Hendershot, J.M.; et al. Dehydration Alters Transcript Levels in the Mosquito Midgut, Likely Facilitating Rapid Rehydration following a Bloodmeal. Insects 2023, 14, 274. https://doi.org/10.3390/insects14030274
Holmes CJ, Brown ES, Sharma D, Warden M, Pathak A, Payton B, Nguyen Q, Spangler A, Sivakumar J, Hendershot JM, et al. Dehydration Alters Transcript Levels in the Mosquito Midgut, Likely Facilitating Rapid Rehydration following a Bloodmeal. Insects. 2023; 14(3):274. https://doi.org/10.3390/insects14030274
Chicago/Turabian StyleHolmes, Christopher J., Elliott S. Brown, Dhriti Sharma, Matthew Warden, Atit Pathak, Blaine Payton, Quynh Nguyen, Austin Spangler, Jaishna Sivakumar, Jacob M. Hendershot, and et al. 2023. "Dehydration Alters Transcript Levels in the Mosquito Midgut, Likely Facilitating Rapid Rehydration following a Bloodmeal" Insects 14, no. 3: 274. https://doi.org/10.3390/insects14030274






