Spodoptera frugiperda (Lepidoptera: Noctuidae) Life Table Comparisons and Gut Microbiome Analysis Reared on Corn Varieties
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Corn Varieties and Larvae Selection for Assessing the Insect Life Table and Microbiome Analysis
2.2. Investigation of Growth Characteristics/Life Table of Larvae in Laboratory-Level
2.3. Host Preference Survey and Damage Score Measurement in Green House (a Semi-Field Level Comparison of Feeding Preferences)
2.4. Inoculation of Larvae to Maize Plants to the Agricultural Fields
2.5. Culture- and Molecular-Based Gut Microbiome Identification
2.6. Nonculture (NGS)-Based Gut Microbiome Analysis
2.7. Ethics Statement
3. Results
3.1. Host Preference and Growth Characteristics by Host/Life Table Analysis
3.2. Evaluation and Assessment of Damage Score of Host Plant Leaves
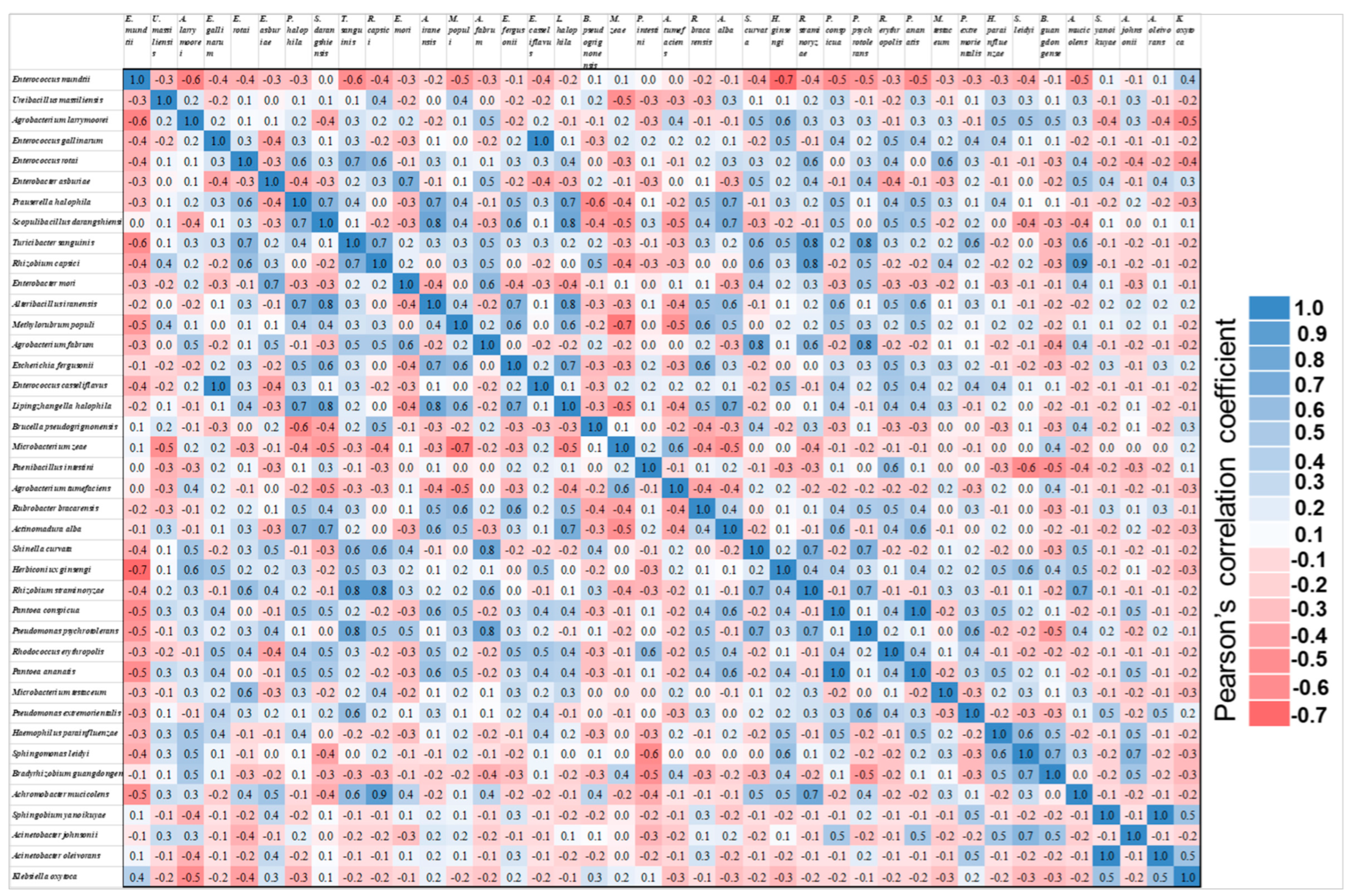
3.3. Microbiome Analysis Based on Corn Varieties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sparks, A.N. A Review of the Biology of the Fall Armyworm. Florida Entomol. 1979, 62, 82–87. [Google Scholar] [CrossRef]
- Day, R.; Abrahams, P.; Bateman, M.; Beale, T.; Clottey, V.; Cock, M.; Colmenarez, Y.; Corniani, N.; Early, R.; Godwin, J.; et al. Fall Armyworm: Impacts and Implications for Africa. Outlooks Pest Manag. 2017, 28, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First Report of Outbreaks of the Fall Armyworm Spodoptera frugiperda (JE Smith)(Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armyworm, F.A.O.F. Map of the Worldwide Spread of Fall Armyworm Since 2016 (as of March 2020), Compiled Using Information from a Range of Sources, Including FAO, International Plant Protection Convention, CABI, the European and Mediterranean Plant Protection Organization, A. 2020. Available online: https://www.eppo.int/ (accessed on 22 March 2023).
- Early, R.; González-Moreno, P.; Murphy, S.T.; Day, R. Forecasting the Global Extent of Invasion of the Cereal Pest Spodoptera frugiperda, the Fall Armyworm. NeoBiota 2018, 40, 25–50. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.N.; Sarker, D.; Pradhan, M.Z.H.; Rashid, M.H. First Report of Occurrence of Fall Armyworm Spodoptera Frugiperda in Bangladesh. Bangladesh J. Entomol. 2018, 28, 97–101. [Google Scholar]
- Sharma, S.K. First Report of Fall Armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae) Incidence in Himachal Pradesh (HP), India. J. Entomol. Res. 2021, 45, 159–164. [Google Scholar] [CrossRef]
- Idrees, A.; Afzal, A.; Qadir, Z.A.; Li, J. Bioassays of Beauveria Bassiana Isolates against the Fall Armyworm, Spodoptera frugiperda. J. Fungi 2022, 8, 717. [Google Scholar] [CrossRef]
- He, L.; Zhao, S.; Gao, X.; Wu, K. Ovipositional Responses of Spodoptera frugiperda on Host Plants Provide a Basis for Using Bt-Transgenic Maize as Trap Crop in China. J. Integr. Agric. 2021, 20, 804–814. [Google Scholar] [CrossRef]
- Wu, M.; Qi, G.; Chen, H.; Ma, J.; Liu, J.; Jiang, Y.; Lee, G.; Otuka, A.; Hu, G. Overseas Immigration of Fall Armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), Invading Korea and Japan in 2019. Insect Sci. 2022, 29, 505–520. [Google Scholar] [CrossRef]
- Lee, G.-S.; Seo, B.Y.; Lee, J.; Kim, H.; Song, J.H.; Lee, W. First Report of the Fall Armyworm, Spodoptera frugiperda (Smith, 1797)(Lepidoptera, Noctuidae), a New Migratory Pest in Korea. Korean J. Appl. Entomol. 2020, 59, 73–78. [Google Scholar]
- Casmuz, A.; Juarez, M.L.; Socias, M.G.; Murua, M.G.; Prieto, S.; Medina, S.; Willink, E.; Gastaminza, G. Review of the Host Plants of Fall Armyworm, Spodoptera Frugiperda (Lepidoptera: Noctuidae). Rev. Soc. Entomol. Argent 2010, 69, 209–231. [Google Scholar]
- Montezano, D.G.; Sosa-Gómez, D.R.; Specht, A.; Roque-Specht, V.F.; Sousa-Silva, J.C.; de Paula-Moraes, S.V.; Peterson, J.A.; Hunt, T.E. Host Plants of Spodoptera Frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Cañas-Hoyos, N.; Márquez, E.J.; Saldamando-Benjumea, C.I. Differentiation of Spodoptera Frugiperda (Lepidoptera: Noctuidae) Corn and Rice Strains from Central Colombia: A Wing Morphometric Approach. Ann. Entomol. Soc. Am. 2014, 107, 575–581. [Google Scholar] [CrossRef]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.-M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagné, N.; Darboux, I. Two Genomes of Highly Polyphagous Lepidopteran Pests (Spodoptera Frugiperda, Noctuidae) with Different Host-Plant Ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, R.N.; Koffi, D.; Agboka, K.; Tounou, K.A.; Banerjee, R.; Jurat-Fuentes, J.L.; Meagher, R.L. Comparative Molecular Analyses of Invasive Fall Armyworm in Togo Reveal Strong Similarities to Populations from the Eastern United States and the Greater Antilles. PLoS ONE 2017, 12, e0181982. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Jiang, X.B.; Ling, Y.; Jiang, T.; Chen, Y.C.; Long, D.; Long, L.P. A Comparative Study on Growth, Development and Reproduction of Mythimna Separata in Four Host Plants. China Plant Prot. 2018, 38, 5–10. [Google Scholar]
- Yin, J.; Cao, Y.Z.; Luo, L.Z.; Hu, Y. Effects of Host Plants on Population Increase of Meadow Moth, Loxostege Sticticalis L. J. Plant Prot. Res. 2004, 31, 173–178. [Google Scholar]
- Idrees, A.; Qadir, Z.A.; Akutse, K.S.; Afzal, A.; Hussain, M.; Islam, W.; Waqas, M.S.; Bamisile, B.S.; Li, J. Effectiveness of Entomopathogenic Fungi on Immature Stages and Feeding Performance of Fall Armyworm, Spodoptera Frugiperda (Lepidoptera: Noctuidae) Larvae. Insects 2021, 12, 1044. [Google Scholar] [CrossRef]
- Altaf, N.; Idrees, A.; Ullah, M.I.; Arshad, M.; Afzal, A.; Afzal, M.; Rizwan, M.; Li, J. Biotic Potential Induced by Different Host Plants in the Fall Armyworm, Spodoptera Frugiperda (Lepidoptera: Noctuidae). Insects 2022, 13, 921. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, J.; He, K.; Zhang, F.; Wang, Z. Potential Invasion of the Crop-Devastating Insect Pest Fall Armyworm Spodoptera Frugiperda to China. Plant Prot. 2018, 44, 1–10. [Google Scholar]
- Da Silva, D.M.; de Bueno, A.F.; Andrade, K.; dos Stecca, C.S.; Neves, P.M.O.J.; de Oliveira, M.C.N. Biology and Nutrition of Spodoptera Frugiperda (Lepidoptera: Noctuidae) Fed on Different Food Sources. Sci. Agric. 2017, 74, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Gressel, J. Microbiome Facilitated Pest Resistance: Potential Problems and Uses. Pest Manag. Sci. 2018, 74, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Engel, P. Mechanisms Underlying Gut Microbiota–Host Interactions in Insects. J. Exp. Biol. 2021, 224, jeb207696. [Google Scholar] [CrossRef]
- Munoz-Benavent, M.; Perez-Cobas, A.E.; Garcia-Ferris, C.; Moya, A.; Latorre, A. Insects’ Potential: Understanding the Functional Role of Their Gut Microbiome. J. Pharm. Biomed. Anal. 2021, 194, 113787. [Google Scholar] [CrossRef]
- Moran, N.A.; Ochman, H.; Hammer, T.J. Evolutionary and Ecological Consequences of Gut Microbial Communities. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 451–475. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.L.; Zhang, V.; Lamberti, L.; Jones, E.W.; Obadia, B.; Korasidis, N.; Gavryushkin, A.; Carlson, J.M.; Beerenwinkel, N.; Ludington, W.B. Microbiome Interactions Shape Host Fitness. Proc. Natl. Acad. Sci. USA 2018, 115, E11951–E11960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.B.; Park, K.-E.; Lee, S.A.; Jang, S.H.; Eo, H.J.; Am Jang, H.; Kim, C.-H.; Ohbayashi, T.; Matsuura, Y.; Kikuchi, Y. Gut Symbiotic Bacteria Stimulate Insect Growth and Egg Production by Modulating Hexamerin and Vitellogenin Gene Expression. Dev. Comp. Immunol. 2017, 69, 12–22. [Google Scholar] [CrossRef]
- Schretter, C.E.; Vielmetter, J.; Bartos, I.; Marka, Z.; Marka, S.; Argade, S.; Mazmanian, S.K. A Gut Microbial Factor Modulates Locomotor Behaviour in Drosophila. Nature 2018, 563, 402–406. [Google Scholar] [CrossRef]
- Li, D.-D.; Li, J.-Y.; Hu, Z.-Q.; Liu, T.-X.; Zhang, S.-Z. Fall Armyworm Gut Bacterial Diversity Associated with Different Developmental Stages, Environmental Habitats, and Diets. Insects 2022, 13, 762. [Google Scholar] [CrossRef]
- Oliveira, N.C.; Rodrigues, P.A.P.; Cônsoli, F.L. Host-Adapted Strains of Spodoptera Frugiperda Hold and Share a Core Microbial Community across the Western Hemisphere. Microb. Ecol. 2022, 15, 1–12. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects–Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. Multiorganismal Insects: Diversity and Function of Resident Microorganisms. Annu. Rev. Entomol. 2015, 60, 17. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Zheng, D.; Zhong, H.; Qin, B.; Gurr, G.M.; Vasseur, L.; Lin, H.; Bai, J.; He, W.; You, M. DNA Sequencing Reveals the Midgut Microbiota of Diamondback Moth, Plutella Xylostella (L.) and a Possible Relationship with Insecticide Resistance. PLoS ONE 2013, 8, e68852. [Google Scholar] [CrossRef]
- Teh, B.-S.; Apel, J.; Shao, Y.; Boland, W. Colonization of the Intestinal Tract of the Polyphagous Pest Spodoptera Littoralis with the GFP-Tagged Indigenous Gut Bacterium Enterococcus mundtii. Front. Microbiol. 2016, 7, 928. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.A.; Murray, B.E. The Rise of the Enterococcus: Beyond Vancomycin Resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Freitak, D.; Vogel, H.; Ping, L.; Shao, Y.; Cordero, E.A.; Andersen, G.; Westermann, M.; Heckel, D.G.; Boland, W. Complexity and Variability of Gut Commensal Microbiota in Polyphagous Lepidopteran Larvae. PLoS ONE 2012, 7, e36978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kwon, M.; Park, K.J.; Maharjanm, R. Monitoring of Four Major Lepidopteran Pests in Korean Cornfields and Management of Helicoverpa armigera. Entomol. Res. 2018, 48, 308–316. [Google Scholar] [CrossRef]
- Jung, J.K.; Seo, B.Y.; Park, C.-G.; Ahn, S.-J.; Kim, J.I.; Cho, J.R. Timing of Diapause Induction and Number of Generations of Helicoverpa armigera (Hüber)(Lepidoptera: Noctuidae) in Suwon, Korea. Korean J. Appl. Entomol. 2015, 54, 383–392. [Google Scholar] [CrossRef]
- Zhang, N.; He, J.; Shen, X.; Sun, C.; Muhammad, A.; Shao, Y. Contribution of Sample Processing to Gut Microbiome Analysis in the Model Lepidoptera, Silkworm Bombyx Mori. Comput. Struct. Biotechnol. J. 2021, 19, 4658–4668. [Google Scholar] [CrossRef] [PubMed]
- Tyrrell, G.J.; Bethune, R.N.; Willey, B.; Low, D.E. Species Identification of Enterococci via Intergenic Ribosomal PCR. J. Clin. Microbiol. 1997, 35, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Berasategui, A.; Salem, H.; Paetz, C.; Santoro, M.; Gershenzon, J.; Kaltenpoth, M.; Schmidt, A. Gut Microbiota of the Pine Weevil Degrades Conifer Diterpenes and Increases Insect Fitness. Mol. Ecol. 2017, 26, 4099–4110. [Google Scholar] [CrossRef]
- Rozadilla, G.; Cabrera, N.A.; Virla, E.G.; Greco, N.M.; McCarthy, C.B. Gut Microbiota of Spodoptera Frugiperda (JE Smith) Larvae as Revealed by Metatranscriptomic Analysis. J. Appl. Entomol. 2020, 144, 351–363. [Google Scholar] [CrossRef]
- Xia, X.; Lan, B.; Tao, X.; Lin, J.; You, M. Characterization of Spodoptera Litura Gut Bacteria and Their Role in Feeding and Growth of the Host. Front. Microbiol. 2020, 11, 1492. [Google Scholar] [CrossRef]
- Funke, M.; Büchler, R.; Mahobia, V.; Schneeberg, A.; Ramm, M.; Boland, W. Rapid Hydrolysis of Quorum-sensing Molecules in the Gut of Lepidopteran Larvae. ChemBioChem 2008, 9, 1953–1959. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Arias-Cordero, E.; Guo, H.; Bartram, S.; Boland, W. In Vivo Pyro-SIP Assessing Active Gut Microbiota of the Cotton Leafworm, Spodoptera littoralis. PLoS ONE 2014, 9, e85948. [Google Scholar] [CrossRef] [Green Version]
- Prem Anand, A.A.; Vennison, S.J.; Sankar, S.G.; Gilwax Prabhu, D.I.; Vasan, P.T.; Raghuraman, T.; Jerome Geoffrey, C.; Vendan, S.E. Isolation and Characterization of Bacteria from the Gut of Bombyx Mori That Degrade Cellulose, Xylan, Pectin and Starch and Their Impact on Digestion. J. Insect Sci. 2010, 10, 107. [Google Scholar]
- Broderick, N.A.; Raffa, K.F.; Goodman, R.M.; Handelsman, J. Census of the Bacterial Community of the Gypsy Moth Larval Midgut by Using Culturing and Culture-Independent Methods. Appl. Environ. Microbiol. 2004, 70, 293–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayatri Priya, N.; Ojha, A.; Kajla, M.K.; Raj, A.; Rajagopal, R. Host Plant Induced Variation in Gut Bacteria of Helicoverpa armigera. PLoS ONE 2012, 7, e30768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Du, K.; Sun, C.; Vimalanathan, A.; Liang, X.; Li, Y.; Wang, B.; Lu, X.; Li, L.; Shao, Y. Gut Bacterial and Fungal Communities of the Domesticated Silkworm (Bombyx mori) and Wild Mulberry-Feeding Relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Sun, B.; Gurr, G.M.; Vasseur, L.; Xue, M.; You, M. Gut Microbiota Mediate Insecticide Resistance in the Diamondback Moth, Plutella xylostella (L.). Front. Microbiol. 2018, 9, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.D.; Mundt, J.O. Enterococci in Insects. Appl. Microbiol. 1972, 24, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, Y.; Wu, W.-M.; Zhao, J.; Jiang, L. Evidence of Polyethylene Biodegradation by Bacterial Strains from the Guts of Plastic-Eating Waxworms. Environ. Sci. Technol. 2014, 48, 13776–13784. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.G.; Milani, C.; De Giori, G.S.; Sesma, F.; Van Sinderen, D.; Ventura, M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, F.; Lu, X. Diversity and Functional Roles of the Gut Microbiota in Lepidopteran Insects. Microorganisms 2022, 10, 1234. [Google Scholar] [CrossRef]
- Grau, T.; Vilcinskas, A.; Joop, G. Probiotic Enterococcus mundtii Isolate Protects the Model Insect Tribolium Castaneum against Bacillus thuringiensis. Front. Microbiol. 2017, 8, 1261. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Luo, S.; Zhou, X. Enterococcus Faecium Regulates Honey Bee Developmental Genes. Int. J. Mol. Sci. 2021, 22, 12105. [Google Scholar] [CrossRef]
- Suriano, S.; Balconi, C.; Valoti, P.; Redaelli, R. Comparison of Total Polyphenols, Profile Anthocyanins, Color Analysis, Carotenoids and Tocols in Pigmented Maize. LWT 2021, 144, 111257. [Google Scholar] [CrossRef]
- Di Lelio, I.; Forni, G.; Magoga, G.; Brunetti, M.; Bruno, D.; Becchimanzi, A.; De Luca, M.G.; Sinno, M.; Barra, E.; Bonelli, M. A Soil Fungus Confers Plant Resistance against a Phytophagous Insect by Disrupting the Symbiotic Role of Its Gut Microbiota. Proc. Natl. Acad. Sci. USA 2023, 120, e2216922120. [Google Scholar] [CrossRef]


| Cultivar ID (Cultivars) | F Value | ||||||
|---|---|---|---|---|---|---|---|
| A (Mebaek 2-ho) | B (Heukjeom 2-ho) | C (Dreamok) | D (Oryun Popcorn) | E (Oryun 2-ho) | F (Meheukchal) | ||
| * Total survival period (days) | † 38.77 ± 17.02 b | 40.40 ± 10.47 ab | 45.50 ± 06.02 a | 41.63 ± 10.09 ab | 40.83 ± 13.51 ab | 44.37 ± 04.89 ab | F (5, 174) = 1.57 |
| p = 0.173 | |||||||
| * Larva period (days) | 19.57 ± 1.48 a | 19.30 ± 1.02 a | 19.23 ± 0.57 a | 17.50 ± 0.90 c | 18.33 ± 0.99 b | 18.16 ± 0.74 b | F (5, 174) = 19.82 |
| p = 0.001 | |||||||
| * Pupa period (days) | 13.26 ±1.14 ab | 13.13 ± 1.10 ab | 13.64 ± 0.78 a | 13.22 ± 1.04 ab | 12.77 ± 1.15 b | 13.10 ± 1.37 ab | F (5, 174) = 2.41 |
| p = 0.039 | |||||||
| * Adult period (days) | 13.26 ± 6.22 ab | 12.68 ± 5.86 ab | 13.50 ± 5.32 a | 15.13 ± 6.14 ab | 15.32 ± 6.16 b | 13.38 ± 4.59 ab | F (5, 174) = 1.16 |
| p = 0.333 | |||||||
| Pupa weight (g) | 0.22 ± 0.03 a | 0.20 ± 0.02 bc | 0.22 ± 0.02 a | 0.20 ± 0.02 c | 0.17 ± 0.02 d | 0.21 ± 0.03 ab | F (5, 174) = 13.66 |
| p = 0.001 | |||||||
| ** Emergency ratio (%) | 93 | 83 | 93 | 82 | 88 | 97 | – |
| ** Pupation ratio (%) | 100 | 83 | 100 | 97 | 97 | 100 | – |
| ** Egg hatching ratio (%) | 57 ab | 32 b | 88 a | 83 a | 73 ab | 73 ab | F (5, 24) = 1.81 |
| p = 0.149 | |||||||
| Cultivars Name | Cultivar ID | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 |
|---|---|---|---|---|---|---|---|---|
| Mebaek 2-ho | A * | † 1.06 ± 0.98 b | 1.78 ± 1.23 b | 2.34 ± 1.36 c | 2.78 ± 1.34 d | 2.88 ± 1.26 d | 3.56 ± 1.48 f | 3.88 ± 1.34 d |
| Heukjeom 2-ho | B | 1.22 ± 1.16 a | 1.91 ± 1.63 a | 2.75 ± 1.68 a | 3.28 ± 1.82 a | 3.50 ± 1.65 b | 4.41 ± 1.56 a | 4.50 ± 1.48 a |
| Dreamok | C | 0.94 ± 0.95 d | 1.75 ± 1.55 c | 2.13 ± 1.52 f | 2.53 ± 1.67 f | 2.69 ± 1.62 f | 3.72 ± 1.80 d | 3.81 ± 1.45 e |
| Oryun popcorn | D | 0.97 ± 1.23 c | 1.63 ± 1.56 d | 2.16 ± 1.72 e | 2.59 ± 1.86 e | 2.18 ± 1.78 e | 3.66 ± 1.93 e | 3.81 ± 1.73 e |
| Oryun 2-ho | E | 0.94 ± 0.88 d | 1.75 ± 1.16 c | 2.28 ± 1.30 d | 2.94 ± 1.37 c | 3.22 ± 1.29 c | 4.00 ± 1.52 c | 4.13 ± 1.58 c |
| Meheukchal | F | 0.81 ± 1.00 e | 1.63 ± 1.26 d | 2.41 ± 1.43 b | 3.00 ± 1.55 b | 3.34 ± 1.23 b | 4.25 ± 1.22 b | 4.34 ± 1.49 b |
| Identify Isolate from FAW Larval Gut | Length (bp) | Coverage (%) | Identity (%) | Ac. No * | Matched to the NCBI (Ac. No.) |
|---|---|---|---|---|---|
| Enterococcus mundtii | 419 | 99 | 99 | OQ184748 | E. mundtii strain DSM 4838 (NZ_CP018061.1) |
| E. casseliflavus | 415 | 100 | 100 | OQ184749 | E. casseliflavus EC20 (NC_020995.1) |
| E. casseliflavus | 415 | 100 | 100 | OQ184750 | E. casseliflavus EC20 (NC_020995.1) |
| E. mundtii | 417 | 100 | 100 | OQ184751 | E. mundtii strain DSM 4838 (NZ_CP018061.1) |
| E. casseliflavus | 415 | 100 | 100 | OQ184752 | E. casseliflavus EC20 (NC_020995.1) |
| E. mundtii | 396 | 100 | 100 | OQ184753 | E. mundtii strain DSM 4838 (NZ_CP018061.1) |
| E. innesii | 415 | 100 | 100 | OQ184754 | E. innesii strain DB-1 (NZ_AP025635.1) |
| E. casseliflavus | 408 | 100 | 100 | OQ184755 | E. casseliflavus EC20 (NC_020995.1) |
| E. casseliflavus | 418 | 99 | 99 | OQ184756 | E. casseliflavus EC20 (NC_020995.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.; Rahman, M.-M.; Han, C.; Shin, J.; Sa, K.J.; Kim, J. Spodoptera frugiperda (Lepidoptera: Noctuidae) Life Table Comparisons and Gut Microbiome Analysis Reared on Corn Varieties. Insects 2023, 14, 358. https://doi.org/10.3390/insects14040358
Jeon J, Rahman M-M, Han C, Shin J, Sa KJ, Kim J. Spodoptera frugiperda (Lepidoptera: Noctuidae) Life Table Comparisons and Gut Microbiome Analysis Reared on Corn Varieties. Insects. 2023; 14(4):358. https://doi.org/10.3390/insects14040358
Chicago/Turabian StyleJeon, Jungwon, Md-Mafizur Rahman, Changhee Han, Jiyeong Shin, Kyu Jin Sa, and Juil Kim. 2023. "Spodoptera frugiperda (Lepidoptera: Noctuidae) Life Table Comparisons and Gut Microbiome Analysis Reared on Corn Varieties" Insects 14, no. 4: 358. https://doi.org/10.3390/insects14040358





