Simple Summary
Black soldier fly (BSF) has considerable potential in the aquaculture industry, not only as a protein source but also as an immunomodulator (a substance that can stimulate or suppress the immune system of an organism). This research is the first to investigate a dietary polysaccharide derived from BSF, called dipterose-BSF, and its potential effect on the immune system of zebrafish. Groups of zebrafish fed on a dipterose-BSF diet were compared with a control group fed on a diet with no dipterose-BSF. Fish in the dipterose-BSF diet group showed increased expression of immune-related genes and decreased expression of stress-related genes compared with the fish in the control group. The optimal concentration of dietary dipterose-BSF needed to enhance the immune status of zebrafish was 0.1 µg/g. A higher concentration (1 µg/g) had no significant effect on the immune system of fish in the dipterose-BSF diet group compared with fish in the control group. Hence, the inclusion of dipterose-BSF in the manufacture of aquafeed should be considered.
Abstract
Dietary management using immunostimulants to protect fish health and prevent bacterial infection is widely practiced. Many insect species possess various bioactive substances that can improve animal health. We previously identified several bioactive polysaccharides derived from insects, including dipterose-BSF from black soldier fly (Hermetia illucens) larvae; this can stimulate innate immunity in mammalian macrophage RAW264.7 cells. However, the effect of dietary dipterose-BSF on the immune system of teleosts remains unclear. Here, we analyzed the immune status of zebrafish (Danio rerio) after 14 days of dietary inclusion of dipterose-BSF (0.01, 0.1, and 1 µg/g), followed by an immersion challenge using Edwardsiella tarda. To identify changes in the transcriptional profile induced by dipterose-BSF, we performed RNA-sequencing analyses of the liver and intestine. Differentially expressed genes were investigated, with both organs showing several upregulated genes, dominated by nuclear factor and tumor necrosis factor family genes. Gene Ontology analysis revealed several terms were significantly higher in the experimental group compared with the control group. Challenge tests suggested that dietary dipterose-BSF had some positive effects on disease resistance in fish, but these effects were not pronounced.
1. Introduction
The most notable and widely farmed insect for animal feed production is the black soldier fly (BSF; Hermetia illucens); it is a non-pest insect [1] and has been widely utilized as a bio-convertor for organic wastes [2,3,4]. BSF larvae contain bioactive substances that can improve animal health; these include antimicrobial peptides (AMPs), which are well-known to be efficient inhibitory compounds against a broad range of pathogens and can be utilized for various purposes [5,6]. Insects can be used in animal feed not only as protein alternatives but also as immunomodulators capable of increasing resistance to diseases [7]. Insects are attracting considerable attention as an animal feed because they are easy to culture, grow rapidly, and they have a short reproductive cycle and a low feed-conversion ratio [8]. Insect meal and its derivatives, such as from silkworm (Bombyx mori) [9,10,11], housefly (Musca domestica) [12,13,14], cricket (Gryllus bimaculatus) [15,16], yellow mealworm (Tenebrio molitor) [17,18,19,20], and black soldier fly (Hermetia illucens) [21,22,23], have been used in various aquaculture commodities to stimulate an immune response, thus increasing stress and disease resistance.
An interesting finding about insects, especially BSF larvae, is that different extraction methods will yield different substances. Diener et al. (2011) [1] and Harlystiarini et al. (2019) [24] found that BSF larvae extracted using methanol had antibacterial activity against Gram-negative bacteria only, with no activity against Gram-positive bacteria. Diener et al. [1] suggested that these differences in susceptibilities between Gram-negative and -positive bacteria may be due to differences in the interactions between the bioactive substance and the bacterial ribosome or the bacterial cell wall. Ali et al. (2019) [25] found that a polysaccharide obtained from BSF larvae using an ethanol extraction method, which they referred to as “dipterose-BSF”, could induce nitric oxide (NO) activity and various proinflammatory cytokines. This indicated that polysaccharides derived from BSF could also have an immunomodulatory effect.
Polysaccharides are reasonably safe, non-toxic, and biodegradable [26], making them excellent for use in livestock and aquaculture. Thus, the use of bioactive polysaccharides could be an appealing alternative to the use of antibiotics and vaccines in aquaculture [27].
The detection of pathogen-associated molecular patterns (PAMPs) by macrophages in lipids, proteins, peptides, nucleic acids, and carbohydrates can activate innate immunity [28,29]. Furthermore, chitin and chitin derivatives, which are composed of N-acetyl-ß-d-glucosamine and are the main component of the exoskeleton of insects, can promote innate immunity to many pathogens as they are also present in pathogenic fungal cell walls [30] and Gram-positive bacteria [31]. The immunomodulatory actions of polysaccharides are generally highly dependent on structural factors, such as their molecular weight, monosaccharide content, and glycosidic bonds [32,33]. Prior research indicates that polysaccharides with a high molecular mass typically exhibit considerably more biological activities compared with the biological activities of polysaccharides with a low molecular mass [34]. This may be due to the greater number of repetitive structures in polysaccharides of high molecular mass, which could interact with receptors or other membrane targets [35].
Here, we extracted and purified dipterose-BSF, we then investigated its bioactive polysaccharide properties by analyzing the NO level induced by mammalian macrophage RAW264.7 cells as a marker for immunomodulatory activity. We conducted an experiment to investigate the immune responses of zebrafish after they received dipterose-BSF as a dietary supplement for 14 days. This was followed by an Edwardsiella tarda challenge test. Furthermore, comprehensive gene expression analysis of zebrafish livers and intestines was conducted using RNA-sequencing (RNA-seq) technology and quantitative real-time polymerase chain reaction (qRT-PCR) analysis. The primary objective of our research was to study the immunomodulatory effects of dipterose-BSF in teleost fish.
2. Materials and Methods
2.1. BSF Larvae Extraction
BSF larvae were obtained from the Research Institute of Environment, Agriculture and Fisheries, Osaka Prefecture, Japan. We used a previously described extraction method [25]. The BSF larvae were autoclaved at 121 °C for 20 min to constrain endogenous enzyme activity; the larvae were then ground into a homogeneous powder. The resulting BSF larvae meal was diluted with ten volumes of ultrapure water and gently shaken at 4 °C for 24 h. The BSF extract was obtained by centrifuging at 10,000× g for 1 h, followed by evaporation in a water bath at 50 °C until one-tenth of its original volume remained, to obtain the concentrated extract.
2.2. Dipterose-BSF Isolation and Purification
Dipterose-BSF was purified according to previously reported methods [25,36,37,38]. To precipitate the dipterose-BSF, 99.5% (v/v) ethanol was added to the BSF extract and gently shaken overnight at 4 °C. The dipterose-BSF was precipitated by centrifugation, washed three times with 80% ethanol, and air-dried inside a draft chamber. The precipitate was diluted in 20 mM Tris-HCl (pH 8.0), shaken overnight at 4 °C, and centrifuged at 10,000× g for 15 min to separate the residual low-molecular-weight substances. Crude dipterose-BSF was obtained once the precipitate had been removed.
Gel-filtration chromatography on a HiPrep 26/10 S-500HR column (GE Healthcare, Chicago, IL, USA) was used for the first stage of the dipterose-BSF purification. The crude dipterose-BSF was loaded into the column, pre-equilibrated with 20 mM Tris-HCl (pH 8.0), and then eluted using the same solution with a flow rate of 1.3 mL/min. The eluted fractions were collected automatically. The sugar content was then evaluated using the phenol–sulfuric acid (PSA) method with glucose as a standard [39] and an evaluation of NO (nitric oxide) activity in macrophage RAW264.7 cells was carried out simultaneously. The PSA- and NO-positive fractions were collected and diluted with the same solution used in the subsequent step.
The positive fractions were then injected into a HiPrep DEAE FF 16/10 column (GE Healthcare, Chicago, IL, USA), pre-equilibrated with 20 mM Tris-HCl (pH 8.0), and eluted at a 2 mL/min flow rate with the same solution. The fractions were collected using a linear gradient of 1 M NaCl of 20%, 40%, and 100% at a 2.0 mL/min flow rate. The eluates were collected automatically, and total sugar and NO assays were performed in the same way as stated earlier. Fractions that exhibited NO activity were pooled and dialyzed using PURELAB ultra-pure water (Elga Veolia, UK) by ten volumes, which was repeated five times. The dialyzed fractions was lyophilized to obtain the dipterose-BSF.
2.3. RAW264.7 Cell Culture
The RAW264.7 cells were provided by the RIKEN Cell Bank (RIKEN BioResource Center, Tsukuba, Japan). Minimal essential medium (MEM) (Life Technologies, Grand Island, NE, USA), supplemented with 10% fetal bovine serum, 0.1 mM non-essential amino acids, 100 U/mL penicillin, and 100 g/mL streptomycin, was used to sustain the cells. The cells were maintained at 37 °C in humidified air containing 5% CO2.
2.4. NO and Sugar Evaluation
The NO production in RAW264.7 cells was measured using a Griess Reagent system kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions. The cells were plated at 1 × 106 cells/well in a 96-well plate, pre-incubated for 90 min, and stimulated for 24 h at 37 °C with various concentrations of dipterose-BSF; 100 ng/mL lipopolysaccharide (LPS) was used as a positive control. The absorbance of the culture medium supernatant was measured at an optical density (OD) of 540 nm using a microplate reader. The quantity of nitrite in the culture media was then determined using an NaNO2 standard curve.
The sugar content was evaluated using the PSA method, as previously described [39]. Briefly, 50 µL of the sample was added to wells in a 96-well plate, then 150 µL of 97% sulfuric acid was added, followed by 30 µL of 5% phenol. The plates were placed in an incubator for 5 min at 90 °C and then the absorbance was measured using a microplate reader at an OD of 490 nm.
2.5. Experimental Fish
The zebrafish (Danio rerio) used in the experiment were obtained from our facility at Ehime University. The experimental fish weighed 380–420 mg, with fork lengths of 32–34 mm; the fish were maintained at 25 ± 2 °C. The feeding regime was at satiation, twice daily (at 09.00 am and 5.00 pm), with a light/dark cycle of 14 h:10 h. All of the fish experiments were conducted in accordance with the Animal Experiments Regulations of Ehime University. The protocol was sanctioned by the Institutional Animal Care and Use Committee (IACUC) of Ehime University (Permit Number: 08K2-1). Surgical procedures (euthanized) and body measurements (anesthetized) were conducted using 2-phenoxyethanol at concentrations of 0.2% and 0.01%, respectively, to minimize suffering.
2.6. Preparation of Dipterose-BSF Diet
The composition of the zebrafish diet is shown in Table 1. Prior to pelletization with a cylindrical granulator (ABV-120L, Akira Kiko, Fukui, Japan), dry materials were thoroughly mixed, supplemented with fish oil, and then blended with water. The pellets were thoroughly air-dried after granulation for 1–2 days at 60 °C to a size of 2–3 mm. The dipterose-BSF diets were created by adding it to final concentrations of 0.01, 0.1, and 1 µg/g. As the quantities of the added dipterose-BSF were very small, their nutritional influence was considered to be insignificant.

Table 1.
Zebrafish feed formulation.
2.7. Zebrafish mRNA Sequencing and Validation Using qRT-PCR
A total of 120 zebrafish were randomly divided into four aquariums (45 × 25 × 25 cm). Each tank received a different dipterose-BSF diet: 0.01, 0.1, or 1 µg/g, or no dipterose-BSF (control), for 14 days. The temperature was maintained at 25 °C with constant air filtering for 24 h, the light/dark cycle was 14 h:10 h, feed was provided to satiation at 09:00 am and 5:00 pm, and 30% of the water was changed from the total volume twice a week. After 14 days of dietary treatment, nine fish from each treatment group were euthanized and dissected to obtain liver and intestine samples for RNA extraction. RNA was isolated using ISOGEN II reagent, following the manufacturer’s protocol. To confirm RNA quality, the concentration and A280/A260 values of the extracted total RNA were measured using a trace spectrophotometer (NanoPhotometer P330, Implen, Munchen, Germany).
RNA-seq analysis was performed using four fish from each treatment, with comparisons made between the control group and the 0.1 µg/g dipterose-BSF treatment group. Samples were adjusted to a total RNA concentration and volume of at least 50 ng and 20 µL, respectively, and complete digestion of genomic DNA was performed using the TURBO DNA-freeTM Kit (Invitrogen, MA, USA). The concentration of the total RNA was measured using a QubitRNA HS Assay Kit (Thermo Fisher Scientific, MA, USA), and the sample qualities were confirmed using a Qsep100 DNA Fragment Analyzer and an RNA R1 Cartridge (BiOptic, Taiwan). The library DNA was set up using a KAPA Stranded mRNAseq kit (KAPA BIO SYSTEMS, MA, USA), according to the manufacturer’s instructions. The concentration of the prepared library DNA solution was assessed using Qubit and dsDNA HS Assay Kits (Thermo Fisher Scientific, MA, USA), and the quality of the library DNA was confirmed using a fragment analyzer and a dsDNA 915 Reagent Kit (Advanced Analytical Technologies, Ames, IA, USA).
Library DNA was cyclized using an MGIEasy Circularization Kit (MGI Tech Co., Ltd., Shenzhen, China) and then synthesized into DNA nanoballs (DNBs) using a DNBSEQ-G400RS High-throughput Sequencing Set (MGI Tech Co., Ltd.). DNB sequences were read using a NextSeq 500 (Illumina, CA, USA) at a sequencing depth of 2 × 76 bp. After eliminating adapter sequences from the obtained sequence data with Cutadapt (ver. 1.9.1) [40], Sickle (ver. 1.33) was used to eliminate bases with a quality score of less than 20 and paired reads with less than 30 bases. The filtered data reads were mapped using STAR (ver. 2.7.11b) to the reference sequence of D. rerio (GRCz11-GCA_000002035.4), to produce a file in the BAM format. The BAM file was indexed using Samtools (ver. 1.19.2) [41]. Reads corresponding to the gene regions of the reference sequence were counted using featureCounts (ver. 3.18) [42]. The TMM-edgeR-TMM pipeline was used to identify differentially expressed genes (DEGs) following normalization using the DEGES normalization technique in TCC (ver. 1.38) [43]. The thresholds for up- and downregulation were set at log2 fold-alterations in genes of 1 or −1 times with a false discovery rate (FDR) of <0.05 when compared with the control group. Multidimensional scaling, heatmap, and volcano plots were generated using the metaseqR2 v1.10 [44] and ggplot2 v3.4.3 [45] packages. ClusterProfiler (version 3.18.1) [46] was used to perform Gene Ontology (GO) enrichment and pathway enrichment analysis (PEA) on the DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/ accessed on 3 January 2024) was also used for PEA.
Seven genes were chosen for quantitative reverse transcription polymerase chain reaction (qRT-PCR) using a PowerTrack SYBR Green Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania) according to the manufacturer’s protocols, with a confirmation assay employed for the same number of sample sets (n = 5 for each group). A High-Capacity cDNA Reverse Transcript Kit (Thermo Fisher Scientific, Vilnius, Lithuania) was used to obtain first-strand complementary DNA (cDNA) from 500 ng of total RNA. The thermocycling process was carried out in 96-well white Multiplate PCR Plates (Bio-Rad Laboratories, Tokyo, Japan), utilizing a qRT-PCR detection system (Bio-Rad Laboratories) with the following conditions: 30 s at 95 °C, then 40 cycles of 5 s at 95 °C and 5 s at 55 °C. Following amplification, melting curve procedures were performed for each gene to verify that only a single product was amplified. The comparative threshold (CT) cycle approach established by Livak and Schmittgen (2001) [47] was used to quantify relative gene expression, with elongation factor 1-alpha (elfa) as an internal reference. Elfa was selected as the housekeeping gene for zebrafish as it has a low degree of variability under a wide range of conditions, e.g., during development, across tissue types, and following chemical treatments [48]. The primers used for qRT-PCR are shown in Table 2.

Table 2.
Primers used in quantitative real-time PCR.
2.8. Challenge Test
For the challenge test, we used a bacterial agent (Edwardsiella tarda) obtained from the kidney of an infected red sea bream (Pagrus major) under Edwardsiellosis, which we obtained from Ainan, Ehime Prefecture, Japan. A preliminary investigation showed that the LD50 of E. tarda in zebrafish was 1 × 107 CFU/mL or an optical density of 0.5 at 660 nm. The E. tarda for the challenge test was prepared in 500 mL of LB medium (inoculation ratio 10 µL/100 mL) and shaken at 170 rpm for 17–18 h in a shaker incubator at 28 °C. The E. tarda culture was then centrifuged at 6000 rpm at 4 °C for 5 min, washed twice with phosphate-buffered saline (PBS) (same volume as the medium), followed by a third addition of PBS and shaking the bottle until the E. tarda formed a homogenous mixture. The immersion for infection ratio was 9:1 between culture water and homogenized E. tarda in PBS for 8 h.
The challenge test was carried out to analyze the immunomodulatory response in zebrafish after the fish had been subjected to 14 days of dipterose-BSF dietary treatment. This involved 40 zebrafish that were randomly divided into four aquariums of dimensions 45 × 25 × 25 cm (n = 10 fish per tank). The challenge test was repeated three times. The aquariums were equipped with a submerged air filtration system, and the fish continued to be fed on the experimental diet. The fish were maintained at 25 ± 2 °C and observed until the day after 100% mortality occurred in the control group.
2.9. Statistical Analysis
The standard error of the mean (SEM) was used to present all of the data. The statistical analyses were carried out using EZR software version 1.64 (https://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmed.html accessed on February 5 2024). The survival rate data were compared using the Kaplan–Meier method, further investigated using the log-rank test followed by Holm’s post hoc test (p < 0.05), while mRNA expression was analyzed using one-way analysis of variance (ANOVA) subjected to Tukey’s post hoc test (p < 0.05).
3. Results
3.1. Transcriptome Analysis Using RNA-Seq after Fish Had Received Dietary Dipterose-BSF
The challenge test showed that the optimum concentration of dipterose-BSF as a feed supplement for enhancing immunity was 0.1 µg/g. Therefore, the challenge test result was included as our first result, ahead of the RNA sequencing results and gene expression results. RNA-seq analysis was conducted to study transcriptome changes in the liver and intestine of zebrafish that had received dietary dipterose-BSF (0.1 µg/g) for 14 days compared with the control group. The raw reads resulted from RNA-seq ranged from 14,811,378 to 16,380,572, with average clean reads between 97.76% and 98.03% (Table S1, Supplementary Material).
The clean reads were mapped using STAR (ver. 2.7.11) and compared with genome assembly GRCz11 (GCF_000002035.6) as a reference, resulting in an average successful mapping rate of 93.41% (Table S2). We used the multidimensional scaling (MDS) method plot to assess the data and check for the possibility of bias and found there was little or no bias, as all samples were clustered in accordance with their respective treatment (Figure S1). Heatmap plots based on normalized counts were generated from the liver (Figure S2) and intestine (Figure S3) samples to analyze the pattern of DEGs.
3.2. Differentially Expressed Genes Induced by Dietary Dipterose-BSF
The DEGs were detected using a threshold value of log2 fold-change ≥ 1 with FDR < 0.05. Analysis of DEGs (TCC package, version 1.38) showed that 748 genes in the liver were differentially expressed between the control group (fed on the basal diet) compared with the group fed on a diet containing 0.1 µg/g dipterose-BSF. The DEGs comprised 369 upregulated genes and 379 downregulated genes. Using the same method, we found 698 DEGs in the intestine, including 381 upregulated and 317 downregulated genes (Figure 1).
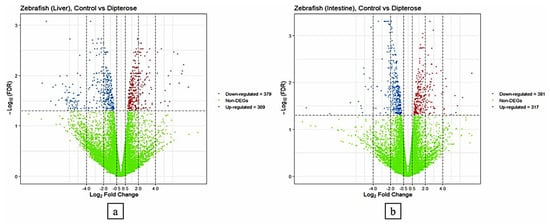
Figure 1.
Transcriptional profiling analysis of the liver and intestine of zebrafish in the control group (fed on the basal diet) compared with the group fed on a diet containing 0.1 µg/g dipterose-BSF. (a) Expression patterns of differentially expressed genes (DEGs) in the livers of zebrafish in the control and dipterose-BSF groups. (b) Expression patterns of DEGs in the intestines of zebrafish in the control and dipterose-BSF groups. The DEGs were analyzed using the TCC package in R software.
3.3. Gene Ontology Enrichment Analysis of DEGs
Next, we used GO enrichment analysis to investigate the molecular mechanisms in the liver and intestines of zebrafish after they received dietary dipterose-BSF. The DEG results from the control and dietary dipterose-BSF groups were then compared by performing an over-representation analysis (ORA) using ClusterProfiler software. In the zebrafish intestine, ORA showed that five molecular functions were enriched (p < 0.05) following the dietary dipterose-BSF treatment: monooxygenase activity, iron ion binding, tubulin binding, heme binding, and cytokine receptor binding (Figure 2).
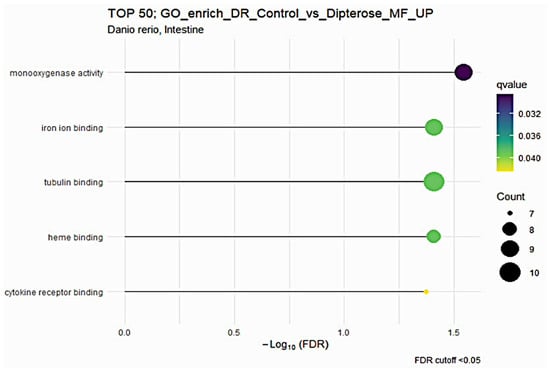
Figure 2.
The Gene Ontology (GO) of molecular function (MF) terms in the zebrafish intestine based on DEGs generated from the comparison between the dipterose-BSF diet group and the control (basal diet) group.
The dipterose-BSF diet impacted more GO terms in the liver of zebrafish compared with in the intestine (p < 0.05). There were twelve GO biological process (BP), four cellular component (CC), and two molecular function (MF) terms (Figure 3). Notably, macroautophagy and processes utilizing autophagic mechanisms were increased, combined with an increase in cytokine receptor binding in the liver, suggesting that dipterose-BSF provides a good foundation for adaptive immunity. This is because during self-degradation or autophagy processes, antigens can be processed then presented by the major histocompatibility complex (MHC) to immune effector cells [49]. Furthermore, one of the functions of cytokines is to regulate the ability of dendritic cells to present antigens and migrate to lymph nodes, which helps to trigger the adaptive immune response [50].
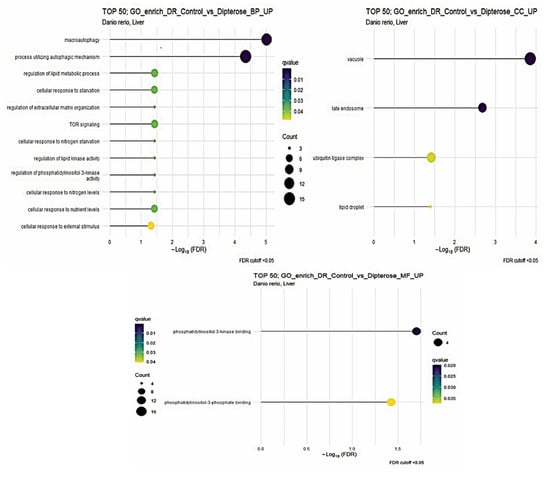
Figure 3.
The Gene Ontology (GO) of biological process (BP), cellular component (CC), and molecular function (MF) terms in the zebrafish liver based on DEGs generated from the comparison between the dipterose-BSF diet group and the control (basal diet) group.
3.4. DEG Signaling Pathway Analysis Using KEGG Pathway Analysis
Evaluation of the signaling pathways of zebrafish liver and intestine samples was carried out using KEGG pathway analysis in ClusterProfiler software. The DEGs compared between the control and treatment groups were processed in the KEGG pathway database (https://www.kegg.jp/ accessed on 3 January 2024) and then analyzed to determine if there were any statistically significant difference terms (p < 0.05). In the liver, five signaling pathways were differentially increased: autophagy (dre04140), nucleotide metabolism (dre01232), PPAR signaling pathway (dre03320), foxO signaling pathway (dre04068), and protein processing in the endoplasmic reticulum (dre04141) (Table 3). At the same time, KEGG pathway analysis of the intestine showed that the ribosome biogenesis in eukaryotes signaling pathway (dre03008) was significantly increased (Table 3).

Table 3.
Significantly enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (p < 0.05) analyzed by ClusterProfiler (ver. 3.18.1) using a comparison of DEGs between the control group and the dietary dipterose-BSF group.
Figure 4 shows a schematic KEGG pathway of protein processing in endoplasmic reticulum (dre04141), which showed that stress-related genes were downregulated, such as Herp, HRD1, as well as HSP family members (HSP40, HSP 70, BiP, and HSP 90).
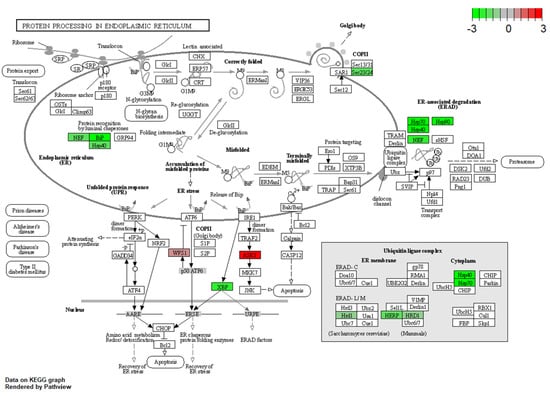
Figure 4.
DEGs relevant to protein processing in endoplasmic reticulum, were generated from the comparison between the dipterose-BSF and the control (basal diet) groups. The green color indicates the downregulated gene, and the red color means the gene was upregulated.
3.5. Immune- and Stress-Related DEG Determination
The determination and categorization of immune- and stress-related DEGs were conducted using research from the literature, the NCBI database, GO annotation, and the KEGG pathway database. Both functional categories were found to be potentially influenced in the zebrafish liver and intestine by dietary dipterose-BSF (Table 4). Further interpretation of the interaction and function of these genes is provided in the discussion.

Table 4.
Full list of differentially expressed immune- and stress-related genes (|log2 fold-change| ≥ 1 and FDR < 0.05) and their expression ratios in the liver and intestine of zebrafish.
3.6. RNA-Seq Validation Using qRT-PCR
RNA-seq results from the comparison of the control and dietary dipterose-BSF groups were validated using qRT-PCR. This verification was performed to ensure the confidence and accuracy of the RNA-seq results. The log2 fold-change obtained from RT-qPCR was compared with the RNA-seq results using Spearman’s correlation (coefficient ρ = 0.929 and p < 0.01) (Figure 5).
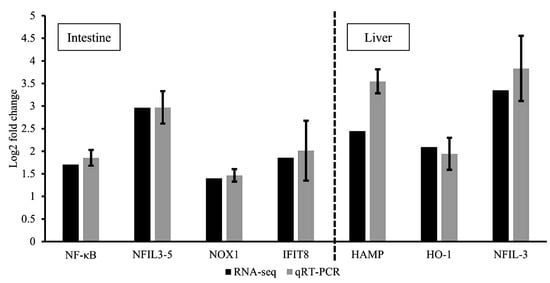
Figure 5.
Validation of RNA-seq results using quantitative RT-PCR. Comparison of relative fold-changes between RNA-seq and qRT-PCR results in zebrafish intestines and liver. NF-κΒ = nuclear factor kappa-light-chain-enhancer of activated B cells; NFIL3-5 = nuclear factor, interleukin 3 regulated, member 5; NOX1 = NADPH oxidase 1; IFIT8 = IFN-induced protein with tetratricopeptide repeats 8; HAMP = hepcidin antimicrobial peptide; HO-1 = heme oxygenase 1; NFIL-3 = nuclear factor, interleukin 3 regulated. Values of qRT-PCR are shown as relative fold-changes between dipterose-BSF treatment and control groups to match the format of the RNA-seq results. Bars indicate the SEM of five samples (n = 5).
3.7. qRT-PCR Analysis of Immune- and Stress-Related Genes
The impacted genes obtained from the zebrafish intestine and liver after dipterose-BSF dietary treatment are having ‘bell-shaped’ dose–response with 0.1 µg/g group as the highest peak. They included nuclear factor kappa-light-chain-enhancer of activated B-cells 2 (NF-κΒ2), nuclear factor interleukin 3, member 5 (NFIL3-5), nicotinamide dinucleotide phosphate oxidase (NOX1), and interferon-induced protein with tetratricopeptide repeats 8 (IFIT8) in the intestine. While Figure 6 shows the relative expression of protectin (CD59), hepcidin antimicrobial peptide (HAMP), heme oxygenase (HO-1), and cells nuclear factor interleukin 3, member 5 (NFIL3-5) in the liver.
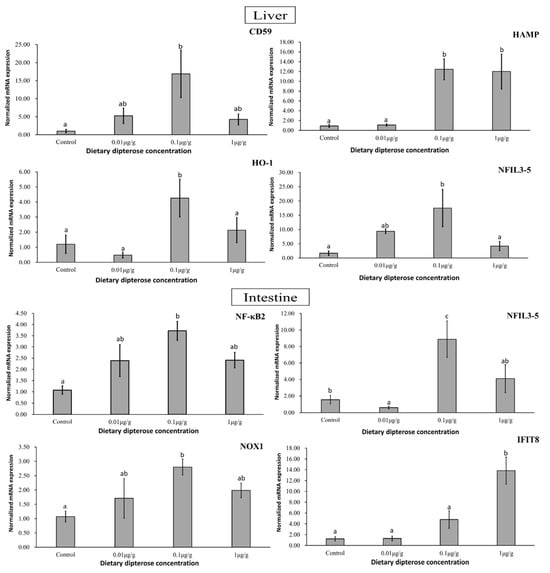
Figure 6.
Expression patterns of immune- and stress-related genes in zebrafish liver and intestine following 14 days of dietary dipterose-BSF treatment using the 2−ΔΔCT method (n = 5). Significance was investigated using a one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Different letters indicate significance (p < 0.05).
3.8. Challenge Test Using E. tarda
The challenge test was carried out to investigate the immune status of zebrafish after they were fed on dipterose-BSF for 14 days and their resistance to infection by pathogen (Figure 7A–C). The zebrafish were challenged with E. tarda and observed until the day after 100% mortality occurred in the control group. The results showed that mortality first appeared on the third day post-challenge in all replicates, with the highest survival rate in the 0.1 µg/g group, with values of 60%, 40%, and 10% shown in replications 1, 2, and 3, respectively. There were no significant differences between the control group and the other treatment groups. The challenge test showed that the optimum concentration of dipterose-BSF as a feed supplement for enhancing immunity was 0.1 µg/g. Therefore, we placed the challenge test result first, ahead of the RNA sequencing results and gene expression results.
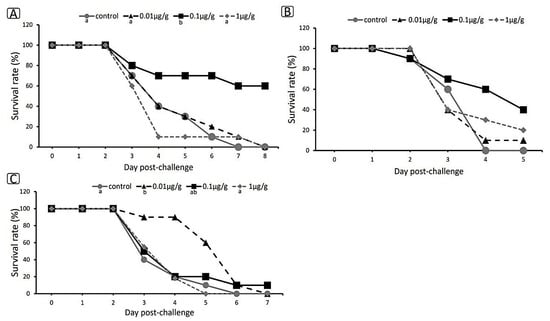
Figure 7.
Survival rate of zebrafish after 14 days of treatment with dietary dipterose-BSF when challenged with E. tarda using the immersion method. (A) Replication 1. (B) Replication 2. (C) Replication 3. Data were compared using the Kaplan–Meier method, statistical significance was obtained with log-rank tests, and the Holm’s method was used for multiple pairwise comparisons (p < 0.05). Small letters indicate significance.
4. Discussion
Previously, we purified multiple bioactive polysaccharides from various insects, including from melon fly larva (Bactrocera cucurbitae), which we named dipterose-BC [36]; Japanese oak pupae (Antheraea yamamai), named silkrose-AY [37]; silkmoth pupae (Bombyx mori), named silkrose-BM [38]; and dipterose-BSF from BSF larvae (H. illucens) [25]. The use of natural substances as immunomodulators in aquaculture has increased considerably, as the use of antibiotics is now widely known to have detrimental effects, both on the environment (leading to the spread of drug-resistance genes) [51] and on humans (resulting in increased severity of infections, which could lead to greater mortality) [52]. According to Ali et al. (2019) [25], dipterose-BSF stimulated Toll-like receptor 2 (TLR2) and TLR4, activating various downstream signaling molecules in RAW264.7 cells, and leading to the expression of numerous proinflammatory cytokines, including interleukin (IL)-1ß, IL-6, and tumor necrosis factor alpha (TNF-α). Dipterose-BSF thus functions more as an immunomodulator than as a bactericidal agent; this would help to reduce the likelihood of resistance genes emerging in the environment.
Dipterose-BSF has practical and economic advantages when used in aquaculture, as BSF can convert low-value organic wastes and by-products into edible sources of protein and fat for fish feed production [53,54]. In the long term, the use of BSF as a fishmeal substitute could reduce commercial feed prices and promote the welfare of fish farmers, especially small-scale fish farmers in low- and middle-income countries who produce freshwater fish.
4.1. Immune- and Stress-Related Genes with Their Respective Receptor Alterations Following Dietary Dipterose-BSF Treatment
Inflammation comprises a complex set of homeostatic mechanisms that involve the neurological, circulatory, and immunological systems and occurs in response to infection or organ injury. If an acute inflammatory response fails to remove a pathogen, the inflammatory process continues and takes on new characteristics [55]. In our study, we found that various immune- and stress-related genes and their respective receptors in the zebrafish liver and intestine were significantly influenced by dietary dipterose-BSF treatment. Pattern recognition receptors (PRRs) are a type of immunological sensor that play critical roles in identifying and responding to conserved patterns found in microorganisms; they include TLRs, RIG-I-like receptors (RLRs), and NOD-like receptors (NLRs), together with their downstream molecules, which have all been identified and characterized in teleosts [56]. Our results showed that TLR/NF-κB was upregulated in both the liver and intestines of zebrafish (Table 4); according to Aoki (2013) [57], this indicates that PAMPs recognize foreign molecules via PRRs. This finding is similar to those reported by Ali and colleagues (2019, 2022) [25,58], who discovered that dipteroses are detected by TLR2 and TLR4, eventually triggering immunomodulatory activities in RAW264.7 cells and Japanese medaka (Oryzias latipes). Furthermore, dipteroses have been shown to increase the expression of NFIL3 and NFIL3-5, which are vital upstream regulators of the NF-κB pathway [59].
An interesting finding in our present study was that the downregulated genes were dominated by antiviral genes, for instance, IRF10, a protein responsible for stimulating antiviral activity in virus-infected cells and also promoting MHC [60]. Presumably, this occurs because dipterose-BSF, which is classified as a polysaccharide, has a structure that resembles the bacterial LPS found in Gram-negative bacteria and can thus activate innate antimicrobial immune responses. Bacterial LPS is a cell wall component of Gram-negative bacteria that can act as a PAMP, enabling host cells to recognize a bacterial invasion and triggering innate immune responses [61].
There is evidence to suggest that adaptive immune responses could also be indirectly affected (upregulated) by dietary dipterose-BSF, such as increased expression of the CD83 molecule, which has been found to be expressed across numerous activated immune cells, including B and T lymphocytes [62]. Fish NF-κB combines T-cell receptor (TCR) and IL-17 signals to modulate ancestral T-cell immune responses to bacterial infection [63].
Regarding stress-related genes, we categorized them based on their association with reactive oxygen species (ROS). The results showed that dietary dipterose-BSF treatment did not cause oxidative stress (Table 4). There were 23 stress-related genes that were downregulated, compared with 12 stress-related genes that were upregulated, due to the increased expression of five serine-related genes. According to Yang and colleagues (2021) [64], serine stimulates glutathione synthesis, which reduces the production of ROS and subsequently suppresses immunological responses, preventing an overactive immune response. Thus, in this study, the increased expression of serine-related genes might act to limit the increases in the immune response caused by dipterose-BSF.
One interesting finding of this study is that dietary dipterose-BSF significantly suppresses the expression of heat shock proteins (HSPs), including HSP40, HSP70, and HSP90 (Figure 4). The accumulation of HSP families is well-known to be associated with the intensity of stress. Thus, these proteins have been regarded as a suitable biomarker for assessing animals’ reactions to environmental and physiological stressors [65]. Taken together, dipterose-BSF might have a stress-reducing effect in teleost, suggesting a promising alternative to tackle stress effects in teleost, such as during transportation or global temperature rise. Further analysis in this regard is needed to confirm these phenomena.
4.2. Selected Immune- and Stress-Related Gene Alterations Following Dietary Dipterose-BSF Treatment Using qRT-PCR
Teleost inflammation processes protect the host from infection and serve as a vital mechanism for the activation of subsequent tissue healing systems [66]. We found that several AMPs, NADPH oxidase, and nuclear factors and their respective receptors were affected significantly in zebrafish liver and intestine after dietary dipterose-BSF treatment, resulting in a ‘bell-shaped’ dose–response, with the 0.1 µg/g group showing the highest peak (Figure 5).
Immune- and stress-related genes whose expression was altered after receiving dietary dipterose-BSF included CD59, HAMP, HO-1, and NFIL-3 (liver), as well as NF-κΒ, NFIL-3, NOX1, and IFIT8 (intestine). Proinflammatory signaling reshapes the milieu within an injury/infection site, stimulates leukocyte recruitment, promotes antimicrobial mechanisms, and leads to the resolution of inflammation [66]. According to Sun et al. (2013) [67], CD59 (also known as protectin) in zebrafish regulates inflammation and protects cell walls and membranes from rupture. This protein is useful throughout all life stages in zebrafish for suppressing cell breakdown caused by viruses. HAMP, known as hepcidin in teleosts, has two different roles: regulating iron metabolism and antimicrobial activity [68]. Furthermore, Neves and colleagues (2015) [69] found that enhanced HAMP1 expression is a response to limit the iron available to pathogens by increasing iron retention and mobilization, while HAMP2 is associated with direct antimicrobial activity. The NF-κΒ signaling pathway is involved in several physiological processes, including inflammation, immunological responses, apoptosis, and cell proliferation and differentiation [70]. In this study, we found that the optimum NF-κΒ expression occurred in the 0.1 µg/g group (p < 0.05). We suspect this is because dipterose-BSF, as a polysaccharide, can activate NF-κΒ. More than 150 distinct stimuli can activate NF-κΒ, including bacterial LPS, proinflammatory cytokines such as TNF-α and IL-1, hormones, and mitogenic substances [71].
In addition to proinflammatory activities, we also found anti-inflammatory responses in zebrafish livers following exposure to dietary dipterose-BSF. We found that heme oxygenase (HO-1) expression was significantly higher in the 0.1 µg/g group. Anti-inflammatory processes in teleosts function to suppress immune responses [72], to prevent excessive inflammation and subsequent tissue damage. Based on this result, we presume that an optimum dose of dietary dipterose-BSF could promote immune homeostasis. In addition, we found that genes related to tissue regeneration, such as NFIL-3 [73] and NOX-1 [74], were also upregulated by the dietary inclusion of dipterose-BSF. NADPH oxidases, or NOXes, are enzymes that play an important role in the production of ROS following injury [75]. Increased levels of NFIL-3 mRNA in myeloid cells could also lead to tissue regeneration post injury and the overall development of the hematopoietic system of zebrafish [73]. Furthermore, zebrafish immune systems attempt to achieve homeostasis conditions, and Table 4 shows that several TNFs are upregulated both in the liver and intestine. According to Li et al. (2021) [76], TNFs are cytokines that play a crucial role in disease pathogenesis and homeostasis. Furthermore, TNFs are one of the most important types of proinflammatory cytokines involved in systemic inflammation and are required for effective innate and adaptive immune responses. These results suggest that dipterose-BSF can not only alter the inflammatory response but also plays a role in maintaining homeostasis and stimulating wound healing, both of which are vital for the survival of fish.
4.3. Challenge Test
We found that the optimum dose of dipterose-BSF to enhance zebrafish immunity was 0.1 µg/g. A higher dose of dipterose-BSF did not provide greater protection against infection (Figure 6). This is because high doses of polysaccharides can have adverse effects on a fish’s immune system/immunosuppression [77] or lead to immune fatigue [78]. In addition, Wu et al. (2023) [79] reported that a high concentration of polysaccharide (laminarin) can cause proteobacteria amplification in the intestine of largemouth bass. Qin et al. (2023) [80] also found that the majority of Bacillus genera, a beneficial bacterium in the intestine, declined to varying degrees following all polysaccharide (laminarin) treatments they tested; however, a low dose of polysaccharide was shown to increase Lactobacillus and Klebsiella abundance in the intestine of spotted seabass (Lateolabrax maculatus), promoting the growth of beneficial bacteria.
As mentioned above, dipterose-BSF stimulates the immune system of zebrafish at the level of gene expression. Although some of our challenge trials showed dipterose-BSF had a significant effect on disease resistance (Figure 7A–C), not all trials showed this, and even those trials did not show sufficiently high survival rates. These results may indicate that dipterose-BSF can increase disease resistance in fish, but not to the same extent that has been observed with silkrose-BM [38].
5. Conclusions
In conclusion, dietary dipterose-BSF, a polysaccharide derived from H. illucens, at a concentration of 0.1 µg/g, effectively increased immune- and stress-related gene expression in zebrafish and reduced E. tarda infection. Transcriptome analysis showed that dipterose-BSF can stimulate many host immune responses to help resist bacterial infection. This study highlights the potential of extracted polysaccharides as a means of disease control in aquaculture. Despite the fact that we thoroughly investigated the effect of dietary dipterose on zebrafish immune responses, further studies should be conducted to investigate the long-term dietary effects, together with other factors such as reproduction, growth, post-harvest quality, and, ultimately, production costs. This will help to determine the optimal usage of dipterose-BSF in commercial aquaculture production.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects15050326/s1, Figure S1: MDS plot verifying biases between C-ZF (control group) as blue circle and BSF-ZF (dipterose-BSF dietary inclusion 0.1 µg/g) as green circle. Samples are shown in columns (n = 4), while DEGs are in rows; Figure S2: The heatmap display of DEGs obtained from zebrafish liver. Samples are shown in columns (n = 4), while DEGs are in rows; Figure S3: The heatmap display of DEGs was obtained from the zebrafish intestine; Table S1: The RNA-seq summary results of raw reads, clean reads, and values of Q20 and Q30 (n = 4); Table S2: The RNA-seq summary table of clean reads and mapping results (n = 4).
Author Contributions
Conceptualization, C.M. and T.M.; methodology, M.F.Z.A.; validation, I.B.B.S. and H.N.; formal analysis, H.N., I.B.B.S., M.F.Z.A. and T.M.; investigation, H.N., I.B.B.S., M.F.Z.A., C.M. and T.M.; data curation, H.N. and I.B.B.S.; Writing—Original draft preparation, I.B.B.S.; Writing—Review and editing, M.F.Z.A. and T.M.; visualization, M.F.Z.A.; supervision, M.F.Z.A., C.M. and T.M.; funding acquisition, T.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the JSPS Kakenhi (18H03960).
Data Availability Statement
The supporting data regarding this research are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank the Research Institute of Environment, Agriculture and Fisheries, Osaka Prefecture, Japan, for providing the BSF larvae.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Choi, W.H.; Yun, J.H.; Chu, J.P.; Chu, K.B. Antibacterial effect of extracts of Hermetia illucens (Diptera: Stratiomyidae) larvae against Gram-negative bacteria. J. Entomol. Res. 2012, 42, 219–226. [Google Scholar] [CrossRef]
- Kim, W.; Bae, S.; Park, K.; Lee, S.; Choi, Y.; Han, S.; Koh, Y. Biochemical characterization of digestive enzymes in the black soldier fly, Hermetia illucens (Diptera: Stratiomyidae). J. Asia Pac. Entomol. 2011, 14, 11–14. [Google Scholar] [CrossRef]
- Diener, S.; Solano, N.M.S.; Gutierrez, F.R.; Zurbrugg, C.; Tockner, K. Biological Treatment of Municipal Organic Waste using Black Soldier Fly Larvae. Waste Biomass Valori. 2011, 2, 357–363. [Google Scholar] [CrossRef]
- Mousavi, S.; Zahedinezhad, S.; Loh, J.Y. A review on insect meals in aquaculture: The immunomodulatory and physiological effects. Int. Aquat. Res. 2020, 12, 100–115. [Google Scholar] [CrossRef]
- Brown, S.E.; Howard, A.; Kasprzak, A.B.; Gordon, K.H.; East, P.D. The discovery and analysis of a diverged family of novel antifungal moricin-like peptides in the wax moth Galler mellonella. Insect Biochem. Mol. Biol. 2008, 38, 201–212. [Google Scholar] [CrossRef]
- Yang, W.; Cheng, T.; Ye, M.; Deng, X.; Yi, H.; Huang, Y.; Tan, X.; Han, D.; Wang, B.; Xiang, Z.; et al. Functional divergence among silkworm antimicrobial peptide paralogs by the activities of recombinant proteins and the induced expression profiles. PLoS ONE 2011, 6, e18109. [Google Scholar] [CrossRef]
- Gasco, L.; Józefiak, A.; Henry, M. Beyond the protein concept: Health aspects of using edible insects on animals. J. Insects Food Feed 2020, 7, 715–741. [Google Scholar] [CrossRef]
- Katayama, N.; Ishikawa, Y.; Takaoki, M.; Yamashita, M.; Nakayama, S.; Kiguchi, K.; Kok, R.; Wada, H.; Mitsuhashi, J.; Space Agriculture Task Force. Entomophagy: A key to space agriculture. ASR 2008, 41, 701–705. [Google Scholar] [CrossRef]
- Sun, Y.; Chang, A.K.; Wen, Z.; Li, Y.; Du, X.; Li, S. Effect of replacing dietary fish meal with silkworm (Bombyx mori L) caterpillar meal on growth and non-specific immunity of sea cucumber Apostichopus japonicus (Selenka). Aquac. Res. 2014, 45, 1246–1252. [Google Scholar] [CrossRef]
- Boonyakida, J.; Nakanishi, T.; Satoh, J.; Shimahara, Y.; Mekata, T.; Park, E.Y. Immunostimulation of shrimp through oral administration of silkworm pupae expressing VP15 against WSSV. Fish Shellfish Immunol. 2022, 128, 157–167. [Google Scholar] [CrossRef]
- Miura, T.; Nishikawa, M.; Otsu, Y.; Ali, M.F.Z.; Hashizume, A.; Miura, C. The effects of silkworm-derived polysaccharide (silkrose) on ectoparasitic infestations in yellowtail (Seriola quinqueradiata) and White Trevally (Pseudocaranx dentex). Fishes 2022, 7, 14. [Google Scholar] [CrossRef]
- Ido, A.; Iwai, T.; Ito, K.; Ohta, T.; Mizushige, T.; Kishida, T.; Miura, C.; Miura, T. Dietary effects of housefly (Musca domestica) (Diptera: Muscidae) pupae on the growth performance and the resistance against bacterial pathogen in red sea bream (Pagrus major) (Perciformes: Sparidae). Appl. Entomol. Zool. 2015, 50, 213–221. [Google Scholar] [CrossRef]
- Li, X.; Rahimnejad, S.; Wang, L.; Lu, K.; Song, K.; Zhang, C. Substituting fish meal with housefly (Musca domestica) maggot meal in diets for bullfrog Rana (Lithobates) catesbeiana: Effects on growth, digestive enzymes activity, antioxidant capacity and gut health. Aquaculture 2019, 499, 295–305. [Google Scholar] [CrossRef]
- Fan, T.; Xiang, J.; Qin, L.; Li, W.; Li, M.; Zou, H.; Song, K.; Wu, S.; Wang, G. Effects of dietary housefly larvae (Musca domestica) on the growth performance, immunity and intestinal microbiota of Chinese soft-shelled turtle (Pelodiscus sinensis). Aquac. Res. 2022, 53, 1862–1872. [Google Scholar] [CrossRef]
- Jeong, S.M.; Khosravi, S.; Mauliasari, I.R.; Lee, B.J.; You, S.G.; Lee, S.M. Nutritional evaluation of cricket, Gryllus bimaculatus, meal as fish meal substitute for oliveflounder, Paralichthys olivaceus, juveniles. J. World Aquac. Soc. 2021, 52, 859–880. [Google Scholar] [CrossRef]
- Fan, K.; Liu, H.; Pei, Z.; Brown, P.B.; Huang, Y. A study of the potential effect of dietary fishmeal replacement with cricket meal (Gryllus bimaculatus) on growth performance, blood health, liver antioxidant activities, intestinal microbiota and immune-related gene expression of juvenile channel catfish. AFST 2023, 295, 115542. [Google Scholar] [CrossRef]
- Henry, M.; Gasco, L.; Piccolo, G.; Fountoulaki, E. Review on the use of insects in the diet of farmed fish: Past and future. AFST 2018, 203, 1–22. [Google Scholar] [CrossRef]
- Ido, A.; Hashizume, A.; Ohta, T.; Takahashi, T.; Miura, C.; Miura, T. Replacement of fish meal by defatted yellow mealworm (Tenebrio molitor) larvae in diet improves growth performance and disease resistance in red seabream (Pargus major). Animals 2019, 9, 100. [Google Scholar] [CrossRef]
- Motte, C.; Rios, A.; Lefebvre, T.; Do, H.; Henry, M.; Jintasataporn, O. Replacing fish meal with defatted insect meal (yellow mealworm Tenebrio molitor) improves the growth and immunity of pacific white shrimp (Litopenaeus vannamei). Animals 2019, 9, 258. [Google Scholar] [CrossRef]
- Shafique, L.; Abdel-Latif, H.M.R.; Hassan, F.-U.; Alagawany, M.; Naiel, M.A.E.; Dawood, M.A.O.; Yilmaz, S.; Liu, Q. The feasibility of using yellow mealworms (Tenebrio molitor): Towards a sustainable aquafeed industry. Animals 2021, 11, 811. [Google Scholar] [CrossRef]
- Xiao, X.; Jin, P.; Zheng, L.; Cai, M.; Yu, Z.; Yu, J.; Zhang, J. Effects of black soldier fly (Hermetia illucens) larvae meal protein as a fishmeal replacement on the growth and immune index of yellow catfish (Pelteobagrus fulvidraco). Aquac. Res. 2018, 49, 1569–1577. [Google Scholar] [CrossRef]
- Chaklader, M.R.; Siddik, M.A.B.; Fotedar, R.; Howieson, J. Insect larvae, Hermetia illucens in poultry by-product meal for barramundi, Lates calcarifer modulates histomorphology, immunity and resistance to Vibrio harveyi. Sci. Rep. 2019, 9, 16703. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.; Abdel-Tawwab, M.; Khalil, R.H.; Metwally, A.A.; Shakweer, M.S.; Ghetas, H.A.; Khallaf, M.A. Black soldier fly (Hermetia illucens) larvae meal in diets of European seabass: Effects on antioxidative capacity, non-specific immunity, transcriptomic responses, and resistance to the challenge with Vibrio Alginolyticus. Fish Shellfish Immunol. 2021, 114, 207–217. [Google Scholar] [CrossRef]
- Harlystiarini, H.; Mutia, R.; Wibawan, I.W.T.; Astuti, D.A. In vitro antibacterial activity of black soldier fly (Hermetia Illucens) larva extracts against gram-negative bacteria. Bul. Peternak. 2019, 43, 125–129. [Google Scholar] [CrossRef]
- Ali, M.F.Z.; Ohta, T.; Ido, A.; Miura, C.; Miura, T. The dipterose of black soldier fly (Hermetia illucens) induces innate immune response through toll-like receptor pathway in mouse macrophage RAW264.7 cells. Biomolecules 2019, 9, 677. [Google Scholar] [CrossRef]
- Li, P.; Wang, F. Polysaccharides: Candidates of promising vaccine adjuvants. Drug Discov. Ther. 2015, 9, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.F.Z.; Kameda, K.; Kondo, F.; Iwai, T.; Kurniawan, R.A.; Ohta, T.; Ido, A.; Takahashi, T.; Miura, C.; Miura, T. Effects of dietary silkrose of Antheraea yamamai on gene expression profiling and disease resistance to Edwardsiella tarda in Japanese medaka (Oryzias latipes). Fish Shellfish Immunol. 2021, 114, 207–217. [Google Scholar] [CrossRef]
- Yan, Z.; Hansson, G.K. Innate immunity, macrophage activation, and atherosclerosis. Immunol. Rev. 2007, 219, 187–203. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Morin-Crini, N.; Lichtfouse, E.; Torri, G.; Crini, G. Fundamentals and Applications of Chitosan. In Sustainable Agriculture Reviews 35; Crini, G., Lichtfouse, E., Eds.; Springer: Cham, Switzerland, 2019; Volume 35, pp. 49–123. [Google Scholar] [CrossRef]
- Marsh, M.B.; Rice, C.D. Development, characterization, and technical applications of a fish lysozyme-specific monoclonal antibody (mAb M24-2). Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, e15–e23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, X.; Duan, Y.; Yang, W.; Zhang, H.; Li, C.; Zhang, J. Structural elucidation and immunostimulatory activity of polysaccharide isolated by subcritical water extraction from Cordyceps militaris. Carbohydr. Polym. 2017, 10, 794–802. [Google Scholar] [CrossRef]
- Bi, S.; Jing, Y.; Zhou, Q.; Hu, X.; Zhu, J.; Guo, Z.; Song, L.; Yu, R. Structural elucidation and immunostimulatory activity of a new polysaccharide from Cordyceps militaris. Food Funct. 2018, 9, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.C.; Shin, H.Y.; Kim, W.J.; Seo, M.S.; Kim, H. Effects of a high-molecular-weight polysaccharides isolated from korean persimmon on the antioxidant, anti-inflammatory, and antiwrinkle activity. Molecules 2021, 26, 1600. [Google Scholar] [CrossRef]
- Rong, Y.; Yang, R.; Yang, Y.; Wen, Y.; Liu, S.; Li, C.; Hu, Z.; Cheng, X.; Li, W. Structural characterization of an active polysaccharide of longan and evaluation of immunological activity. Carbohydr. Polym. 2019, 213, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Ido, A.; Kusano, K.; Miura, C.; Miura, T. A novel polysaccharide in insects activates the innate immune system in mouse macrophage RAW264 cells. PLoS ONE 2014, 9, e114823. [Google Scholar] [CrossRef]
- Ohta, T.; Kusano, K.; Ido, A.; Miura, C.; Miura, T. Silkrose: A novel acidic polysaccharide from the silkmoth that canstimulate the innate immune response. Carbohydr. Polym. 2016, 136, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.F.Z.; Yasin, I.A.; Ohta, T.; Hashizume, A.; Ido, A.; Takahashi, T.; Miura, C.; Miura, T. The silkrose of Bombyx mori effectively prevent vibriosis in penaeid prawns via the activation of innate immunity. Sci. Rep. 2018, 8, 8836. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from highthroughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Moulos, P.; Hatzis, P. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleid Acid Res. 2015, 43, e25. [Google Scholar] [CrossRef]
- Wilkinson, L. ggplot2: Elegant graphic for data analysis by WICKHAM, H. Biometerics 2011, 67, 678–679. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCΤ method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- McCurley, A.T.; Callard, G.V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008, 9, 102. [Google Scholar] [CrossRef]
- Johnstone, C.; Chavez-Pozo, E. Antigen Presentation and Autophagy in Teleost Adaptive Immunity. Int. J. Mol. Sci. 2022, 23, 4899. [Google Scholar] [CrossRef]
- Wang, T.; Secombes, C.J. The cytokine networks of adaptive immunity in fish. Fish Shellfish Immunol. 2013, 35, 1703–1718. [Google Scholar] [CrossRef]
- Xia, J.; Ge, C.; Yao, H. Antimicrobial peptides from black soldier fly (Hermetia illucens) as potential antimicrobial factors representing an alternative to antibiotics in livestock farming. Animals 2021, 11, 1937. [Google Scholar] [CrossRef]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Čičková, H.; Newton, G.L.; Lacy, R.C.; Kozánek, M. The use of fly larvae for organic waste treatment. Waste Manag. 2015, 35, 68–80. [Google Scholar] [CrossRef]
- Meneguz, M.; Schiavone, A.; Gai, F.; Dama, A.; Lussiana, C.; Renna, M.; Gasco, L. Effect of rearing substrate on growth performance, waste reduction efficiency and chemical composition of black soldier fly (Hermetia illucens) larvae. J. Sci. Food Agric. 2018, 98, 5776–5784. [Google Scholar] [CrossRef] [PubMed]
- Dezfuli, B.S.; Lorenzoni, M.; Carosi, A.; Giari, L.; Bosi, G. Teleost innate immunity, an intricate game between immune cells and parasites of fish organs: Who wins, who loses. Front. Immunol. 2023, 14, 1250835. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Su, J. Progresses on three pattern recognition receptor families (TLRs, RLRs and NLRs) in teleost. Dev. Comp. Immunol. 2021, 122, 104131. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Hikima, J.I.; Hwang, S.D.; Jung, T.S. Innate immunity of finfish: Primordial conservation and function of viral RNA sensors in teleosts. Fish Shellfish Immunol. 2013, 35, 1689–1702. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.F.Z.; Nakahara, S.; Otsu, Y.; Ido, A.; Miura, C.; Miura, T. Effects of functional polysaccharide from silkworm as an immunostimulant on transcriptional profiling and disease resistance in fish. J. Insects Food Feed 2022, 8, 1221–1233. [Google Scholar] [CrossRef]
- Zhuo, H.; Liu, J. Nuclear factor interleukin 3 (NFIL3) participates in regulation of the NF-κB-mediated inflammation and antioxidant system in Litopenaeus vannamei under ammonia-N stress. Fish Shellfish Immunol. 2022, 131, 1192–1205. [Google Scholar] [CrossRef]
- Nehyba, J.; Hrdličková, R.; Burnside, J.; Bose, H.R., Jr. A novel interferon regulatory factor (IRF), IRF-10, has a unique role in immune defense and is induced by the v-rel oncoprotein. Mol. Cell. Biol. 2022, 22, 3942–3957. [Google Scholar] [CrossRef]
- Matsuura, M. Structural modifications of bacterial lipopolysaccharide that facilitate Gram-negative bacteria evasion of host innate immunity. Front. Immunol. 2013, 4, 109. [Google Scholar] [CrossRef]
- Grosche, L.; Knippertz, I.; König, C.; Royzman, D.; Wild, A.B.; Zinser, E.; Sticht, H.; Muller, Y.A.; Steinkasserer, A.; Lechmann, M. The CD83 molecule—An important immune checkpoint. Front. Immunol. 2020, 11, 721. [Google Scholar] [CrossRef]
- Wei, X.; Li, C.; Zhang, Y.; Li, K.; Li, J.; Ai, K.; Li, K.; Zhang, J.; Yang, J. Fish NF-κB couples TCR and IL-17 signals to regulate ancestral T-cell immune response against bacterial infection. FASEB J. 2021, 35, e21457. [Google Scholar] [CrossRef]
- Yang, D.-X.; Yang, M.-J.; Yin, Y.; Kou, T.-S.; Peng, L.-T.; Chen, Z.-G.; Zheng, J.; Peng, B. Serine metabolism tunes immune responses to promote Oreochromis niloticus survival upon Edwardsiella tarda infection. mSystems 2021, 6, e00426-21. [Google Scholar] [CrossRef]
- Hallare, A.V.; Köhler, H.-R.; Triebskorn, R. Developmental toxicity and stress protein responses in zebrafish embryos after exposure to diclofenac and its solvent, DMSO. Chemosphere 2004, 56, 659–666. [Google Scholar] [CrossRef]
- Soliman, A.M.; Barreda, D.R. The acute inflammatory response of teleost fish. Dev. Comp. Immunol. 2023, 146, 104731. [Google Scholar] [CrossRef]
- Sun, C.; Wu, J.; Liu, S.; Li, H.; Zhang, S. Zebrafish CD59 has both bacterial-binding and inhibiting activities. Dev. Comp. Immunol. 2013, 41, 178–188. [Google Scholar] [CrossRef]
- Rodrigues, P.N.S.; Vázquez-Dorado, S.; Neves, J.V.; Wilson, J.M. Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev. Comp. Immunol. 2006, 30, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Caldas, C.; Vieira, I.; Ramos, M.F.; Rodrigues, P.N.S. Multiple hepcidins in a teleost fish, Dicentrarchus labrax: Different hepcidins for different roles. J. Immunol. 2015, 195, 2696–2709. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.G.; Tergaonkar, V.; Ng, J.K.; Dubova, I.; Izpisua-Belmonte, J.C.; Verma, I.M. Characterization of NF-κΒ/IκΒ proteins in zebra fish and their involvement in notochord development. Mol. Cell. Biol. 2004, 24, 5257–5268. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-κΒ transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef]
- Wattrus, S.J.; Zon, L.I. Blood in the water: Recent uses of zebrafish to study myeloid biology. Curr. Opin. Hematol. 2021, 28, 43–49. [Google Scholar] [CrossRef]
- Weaver, C.J.; Leung, Y.F.; Suter, D.M. Expression dynamics of NADPH oxidases during early zebrafish development. J. Comp. Neurol. 2016, 524, 2130–2141. [Google Scholar] [CrossRef]
- Chopra, K.; Folkmanaitė, M.; Stockdale, L.; Shathish, V.; Ishibashi, S.; Bergin, R.; Amich, J.; Amaya, E. Duox is the primary NADPH oxidase responsible for ROS production during adult caudal fin regeneration in zebrafish. iScience 2023, 26, 106147. [Google Scholar] [CrossRef]
- Li, K.; Qiu, H.; Yan, J.; Shen, X.; Wei, X.; Duan, M.; Yang, J. The involvement of TNF-α and TNF-β as proinflammatory cytokines in lymphocyte-mediated adaptive immunity of Nile tilapia by initiating apoptosis. Dev. Comp. Immunol. 2021, 115, 103884. [Google Scholar] [CrossRef]
- Machuca, C.; Martínez, Y.M.; Becerril, M.R.; Angulo, C. Yeast β-glucans as fish immunomodulators: A review. Animals 2022, 12, 2154. [Google Scholar] [CrossRef]
- Yu, W.; Yang, Y.; Zhou, Q.; Huang, X.; Huang, Z.; Li, T.; Wu, Q.; Zhou, C.; Ma, Z.; Lin, H. Effects of dietary Astragalus polysaccharides on growth, health and resistance to Vibrio harveyi of Lates calcarifer. Int. J. Biol. Macromol. 2022, 207, 850–858. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, Y.; Qian, S.; Zhang, W.; Huang, M.; Yang, S.; Fei, H. An evaluation of laminarin additive in the diets of juvenile largemouth bass (Micropterus salmoides): Growth, antioxidant capacity, immune response and intestinal microbiota. Animals 2023, 13, 459. [Google Scholar] [CrossRef]
- Qin, H.; Long, Z.; Ma, J.; Kong, L.; Lin, H.; Zhou, S.; Lin, Y.; Huang, Z.; Liu, L.; Li, Z. Growth performance, digestive capacity and intestinal health of juvenile spotted seabass (Lateolabrax maculatus) fed dietary laminarin supplement. Front. Mar. Sci. 2023, 10, 1242175. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).