Label-Free Differential Proteomics and Quantification of Exoenzymes from Isolates of the Entomopathogenic Fungus Beauveria bassiana
Abstract
:1. Introduction
2. Experimental Section
2.1. B. bassiana Isolation and Maintenance
2.2. Liquid Growth
2.3. Enzyme Activity
2.4. Proteomics Sample Preparation
2.5. Differential Label-Free Quantitative and Qualitative Proteomics
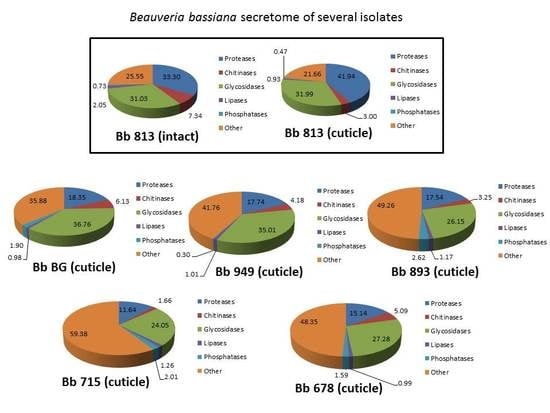
3. Results
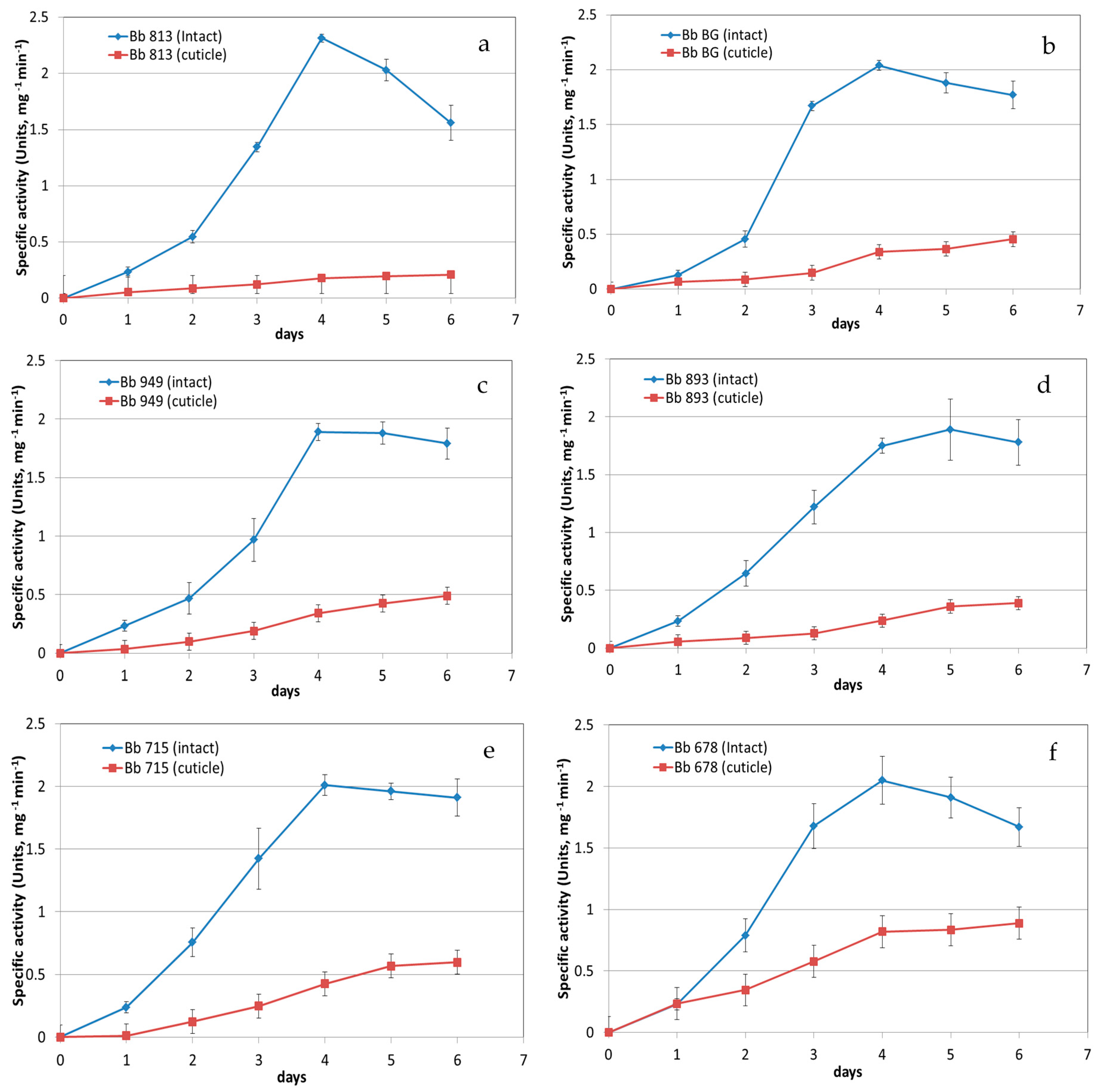
3.1. Protease Activity Measurement
3.2. Proteomics Identification of B. bassiana Exoenzymes
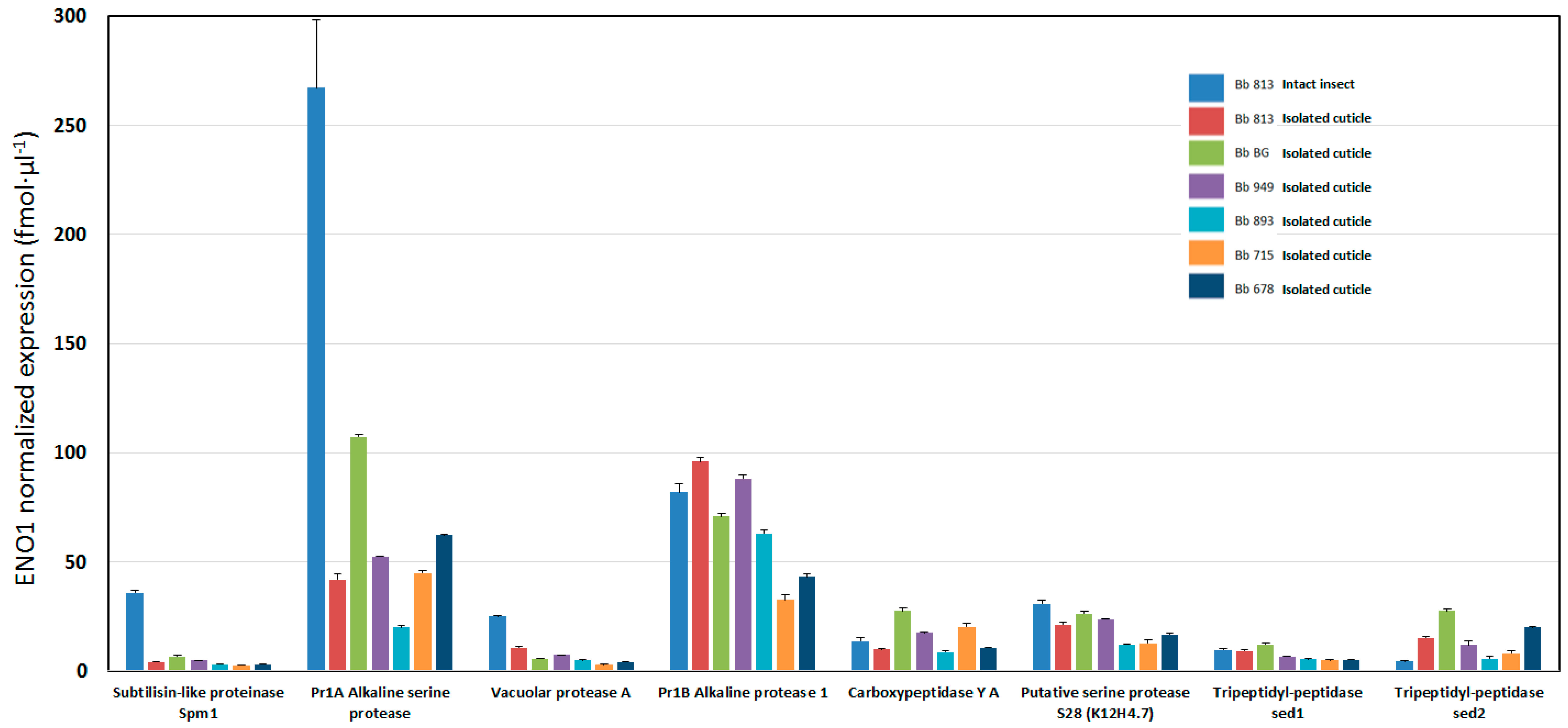
3.3. Qualitative and Quantitative Label-Free Differential Proteomics of B. bassiana Exoenzymes among the Five Isolates
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Barelli, L.; Moonjely, S.; Behie, S.W.; Bidochka, M.J. Fungi with multifunctional lifestyles: Endophytic insect pathogenic fungi. Plant Mol. Biol. 2016, 90, 657–664. [Google Scholar] [CrossRef] [PubMed]
- Clarkson, J.M.; Charnley, A.K. New insights into the mechanisms of fungal pathogenesis in insects. Trends Microbiol. 1996, 4, 197–203. [Google Scholar] [CrossRef]
- Ferron, P. Pest control by the fungi Beauveria and Metarhizium. In Microbial Control of Pests and Plant Diseases 1970–1980; Burges, H.D., Ed.; Academic Press: New York, NY, USA, 1981; pp. 465–482. [Google Scholar]
- Hackman, R.H. Cuticle: Biochemistry. In Biology of the Integument; Bereiter-Hahn, J., Mateltsy, A.G., Richards, K.S., Eds.; Springer: Berlin, Germany, 1984; pp. 583–610. [Google Scholar]
- Anderson, S.O.; Hojrup, P.; Roepstorff, P. Insect cuticular proteins. Insect Biochem. Mol. Biol. 1995, 25, 153–176. [Google Scholar] [CrossRef]
- Mills, R.R.; Fox, F.R. The sclerotization process by the American cockroach: Contribution of melanin. Insect Biochem. 1972, 2, 23–28. [Google Scholar] [CrossRef]
- Kramer, S.; Wigglesworth, V.B. The outer layers of the cuticle in the cockroach Periplaneta americana and the function of the oenocytes. J. Cell Sci. 1950, 91, 63–72. [Google Scholar]
- St Leger, R.J.; Cooper, R.M. Cuticle degrading entomopathogenic fungi: Cuticle degradation in vitro by enzymes from entomopathogenes. J. Invertebr. Pathol. 1986, 41, 167–177. [Google Scholar] [CrossRef]
- St leger, R.J.; Cooper, R.M.; Charnley, A.K. Production of cuticle-degrading enzymes by the entomopathogen Metarhizium anisopliae during infection of cuticles from Calliphora vomitoria and Manduca sexta. J. Gen. Microbiol. 1987, 133, 1371–1382. [Google Scholar] [CrossRef]
- Samsinakova, A.; Misikova, S.; Leopold, J. Action of enzymatic systems of Beauveria bassiana on the cuticle of the greater wax moth (Galleria mellonella). J. Invertebr. Pathol. 1971, 18, 322–330. [Google Scholar] [CrossRef]
- Smith, R.J.; Pekrul, S.; Grula, E.A. Requirement for sequential enzymatic activities for penetration of the integument of the corn earworm (Heliothis zea). J. Invertebr. Pathol. 1981, 38, 335–344. [Google Scholar] [CrossRef]
- Kaur, G.; Padmaja, V. Relationships among activities of extracellular enzyme production and virulence against Helicoverpa armigera in Beauveria bassiana. J. Basic Microbiol. 2009, 49, 264–74. [Google Scholar] [CrossRef] [PubMed]
- Beys-da-Silva, W.O.; Santi, L.; Berger, M.; Calzolari, D.; Passos, D.O.; Guimarães, J.A.; Moresco, J.J.; Yates, J.R. Secretome of the biocontrol agent Metarhizium anisopliae induced by the cuticle of the cotton pest Dysdercus peruvianus reveals new insights into infection. J. Proteome Res. 2014, 13, 2282–2296. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Guo, L.; Shi, H.; Mijit, M.; Qiu, D. Bioassay and enzymatic comparison of six entomopathogenic fungal isolates for virulence or toxicity against green peach aphids Myzus persicae. Afr. J. Biotechnol. 2007, 11, 14193–14203. [Google Scholar] [CrossRef]
- Vici, A.C.; da Cruz, A.F.; Facchini, F.D.; de Carvalho, C.C.; Pereira, M.G.; Fonseca-Maldonado, R.; Ward, R.J.; Pessela, B.C.; Fernandez-Lorente, G.; Torres, F.A.; et al. Beauveria bassiana Lipase A expressed in Komagataella (Pichia) pastoris with potential for biodiesel catalysis. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Jin, K.; Ying, S.H.; Zhang, Y.; Xiao, G.; Shang, Y.; Duan, Z.; Hu, X.; Xie, X.Q.; Zhou, G.; et al. Genome Sequencing and Comparative Transcriptomics of the Model Entomopathogenic Fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; St Leger, R.J.; Wang, C. Advances in Genomics of Entomopathogenic Fungi. Adv. Genet. 2016, 94, 251–285. [Google Scholar]
- Lee, S.B.; Milgroom, M.G.; Taylor, J.W. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genet. Newsl. 1988, 35, 23–24. [Google Scholar]
- Bratton, A.C.; Marshall, E.K. A New Coupling reaction for Sulfanilamide determination. J. Biol. Chem. 1939, 128, 537–550. [Google Scholar]
- Bradford, M.M. A rapid and sensitive for the quantitation of microgram quantitites of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1972, 72, 248–254. [Google Scholar] [CrossRef]
- Dhar, P.; Kaur, G. Cuticle-Degrading Proteases Produced by Metarhizium anisopliae and Their Induction in Different Media. Indian J. Microbiol. 2010, 50, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Dhar, P.; Kaur, G. Production of cuticle—Degrading proteases by Beauveria bassiana and their induction in different media. Afr. J. Biochem. Res. 2010, 4, 65–72. [Google Scholar]
- Wang, C.; St Leger, R.J. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot. Cell 2005, 4, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Noronha, E.F.; Miller, R.N.; Costa, F.T.; Pereira, C.D.; Mehta, A.; Caldas, R.A.; Franco, O.L. Proteomic analysis of Metarhizium anisopliae secretion in the presence of the insect pest Callosobruchus maculatus. Microbiology 2008, 154, 3766–3774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Feng, M.G.; Fan, Y.H.; Luo, Z.B.; Yang, X.Y.; Wu, D.; Pei, Y. A cuticle-degrading protease (CDEP-1) of Beauveria bassiana enhances virulence. Biocontrol Sci. Technol. 2008, 18, 551–563. [Google Scholar] [CrossRef]
- St Leger, R.J.; Joshi, L.; Bidochka, M.J.; Roberts, D.W. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 1996, 93, 6349–6354. [Google Scholar] [CrossRef] [PubMed]
- Leão, M.P.; Tiago, P.V.; Andreote, F.D.; de Araújo, W.L.; de Oliveira, N.T. Differential expression of the pr1A gene in Metarhizium anisopliae and Metarhizium acridum across different culture conditions and during pathogenesis. Genet. Mol. Biol. 2015, 38, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Feng, J.; Fan, Y.; Zhang, Y.; Bidochka, M.J.; Leger, R.J.; Pei, Y. Expressing a fusion protein with protease and chitinase activities increases the virulence of the insect pathogen Beauveria bassiana. J. Invertebr. Pathol. 2009, 102, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Xie, M.; Zhang, X.L.; Peng, D.L.; Yu, W.B.; Li, Q.; Li, Q.; Zhao, J.J.; Zhang, Z.R. Establishment of the PEG-mediated protoplast transformation for Lecanicillium lecanii and development of virulence-enhanced strains against Aphis gossypii. Pest Manag. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Typas, M.A.; Butt, T.M. Detection and characterisation of pr1 virulent gene deficiencies in the insect pathogenic fungus Metarhizium anisopliae. FEMS Microbiol. Lett. 2002, 213, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Santi, L.; Silva, W.O.; Pinto, A.F.; Schrank, A.; Vainstein, M.H. Metarhizium anisopliae host-pathogen interaction: Differential immunoproteomics reveals proteins involved in the infection process of arthropods. Fungal Biol. 2010, 114, 312–319. [Google Scholar] [CrossRef] [PubMed]
- St Leger, R.J.; Charnley, A.K.; Cooper, R.M. Characterization of cuticle-degrading proteases produced by the entomopathogen Metarhizium anisopliae. Arch. Biochem. Biophys. 1987, 253, 221–232. [Google Scholar] [CrossRef]
- Pedrini, N.; Ortiz-Urquiza, A.; Huarte-Bonnet, C.; Zhang, S.; Keyhani, N.O. Targeting of insect epicuticular lipids by the entomopathogenic fungus Beauveria bassiana: Hydrocarbon oxidation within the context of a host-pathogen interaction. Front. Microbiol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Feng, M. Reliability of extracellular protease and lipase activities of Beauveria bassiana isolates used as their virulence indices. Acta Microb. Sin. 1998, 38, 461–467. [Google Scholar]
- Mustafa, U.; Kaur, G. Extracellular enzyme production in Metarhizium anisopliae isolates. Folia Microbiol. (Praha) 2009, 54, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.; Xie, X.Q.; Shang, Y.; St Leger, R.J.; Zhao, G.P.; Wang, C.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012. [Google Scholar] [CrossRef] [PubMed]
- Havukkala, I.; Mitamura, C.; Hara, S.; Hirayae, K.; Nishizawa, Y.; Hibi, T. Induction and Purification of Beauveria bassiana Chitinolytic Enzymes. J. Invertebr. Pathol. 1993, 61, 97–102. [Google Scholar] [CrossRef]
- Dhar, P.; Kaur, G. Effects of carbon and nitrogen sources on the induction and repression of chitinase enzyme from Beauveria bassiana isolates. Afr. J. Biotechol. 2010, 9, 8092–8099. [Google Scholar]
- Fang, W.; Leng, B.; Xiao, Y.; Jin, K.; Ma, J.; Fan, Y.; Feng, J.; Yang, X.; Zhang, Y.; Pei, Y. Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl. Environ. Microbiol. 2005, 71, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Joris, B.; Englebert, S.; Chu, C.P.; Kariyama, R.; Daneo-Moore, L.; Shockman, G.D.; Ghuysen, J.M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 1992, 70, 257–264. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van der Krol, S.; Shibuya, N.; Joosten, M.H.; Thomma, B.P. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Saleem-Batcha, R.; Kombrink, A.; Hansen, G.; Valkenburg, D.J.; Thomma, B.P.; Mesters, J.R. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2013, 2, e00790. [Google Scholar] [CrossRef] [PubMed]
- Kombrink, A.; Bart, P.H.J.; Thomma, B.P.H.J. LysM Effectors: Secreted Proteins Supporting Fungal Life. PLoS Pathog. 2013, 9, e1003769. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Lahrmann, U.; Güldener, U.; Langen, G.; Pfiffi, S.; Biedenkopf, D.; Wong, P.; Samans, B.; Grimm, C.; Basiewicz, M.; et al. Endophytic Life Strategies Decoded by Genome and Transcriptome Analyses of the Mutualistic Root Symbiont Piriformospora indica. PLoS Pathog. 2011, 7, e1002290. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Rafiqi, M.; Jelonek, L.; Akum, N.F.; Zhang, F.; Kogel, K.H. Effector candidates in the secretome of Piriformospora indica, a ubiquitous plant-associated fungus. Front. Plant Sci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.L.; St Leger, R.J. Insect Immunity to Entomopathogenic Fungi. Adv. Genet. 2016, 94, 251–285. [Google Scholar] [PubMed]
- Jiang, R.; Kim, E.H.; Gong, J.H.; Kwon, H.M.; Kim, C.H.; Ryu, K.H.; Park, J.W.; Kurokawa, K.; Zhang, J.; Gubb, D.; et al. Three pairs of protease-serpin complexes cooperatively regulate the insect innate immune responses. J. Biol. Chem. 2009, 284, 35652–35658. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Kanost, M.R. Characterization and functional analysis of 12 naturally occurring reactive site variants of Serpin-1 from Manduca sexta. J. Biol. Chem. 1997, 272, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Joop, G.; Vilcinskas, A. Coevolution of parasitic fungi and insect hosts. Zoology 2016, 119, 350–358. [Google Scholar] [CrossRef] [PubMed]
| Accession | Description | mW (Da) | Peptides | Coverage (%) |
|---|---|---|---|---|
| A0A0A2W0R3 | Pr1A Alkaline serine protease | 38,793 | 12 | 38.2586 |
| A0A0A2VW01 | Pr1B Alkaline protease 1 | 41,528 | 11 | 27.8481 |
| A0A0A2VRL8 | l-ascorbate peroxidase | 101,456 | 8 | 10.9959 |
| A0A0A2VJ73 | Alpha galactosidase | 57,651 | 7 | 12.6629 |
| A0A0A2VDM1 | Putative glycosidase crf1 | 38,406 | 6 | 17.4142 |
| A0A0A2VIV9 | Isochorismatase-like (Cysteine hydrolases) protein | 25,783 | 5 | 18.8034 |
| A0A0A2VJP2 | ATP dependent RNA helicase glh 2 | 51,230 | 5 | 11.8068 |
| A0A0A2VQS8 | Cell_wall_mannoprotein_1 | 19,361 | 5 | 22.4599 |
| A0A0A2V7C2 | Beta-1,3 exoglucanase (Carbohydrate-binding WSC domain) | 18,985 | 4 | 21.5909 |
| A0A0A2V928 | Catalase peroxidase | 87,240 | 4 | 3.7641 |
| A0A0A2VPJ3 | Extracellular aldono-Lactonase | 41,232 | 4 | 10.4859 |
| A0A0A2VSY1 | Subtilisin like proteinase Spm1 | 56,909 | 4 | 6.5789 |
| A0A0A2VUK5 | Endo chitosanase | 31,664 | 4 | 14.6667 |
| A0A0A2V8V1 | Vacuolar protease A | 42,377 | 3 | 7.0886 |
| A0A0A2VHX5 | Six-hairpin glycosidase GH13 | 98,237 | 3 | 4.6804 |
| A0A0A2VL54 | Autolysin (Lysine Motif, LysM domain-containing protein) | 44,154 | 3 | 6.5375 |
| A0A0A2VR76 | Carboxypeptidase Y A | 97,773 | 3 | 2.4719 |
| A0A0A2V6S0 | LysM motifs (double) protein | 49,033 | 2 | 3.9735 |
| A0A0A2VB74 | Endo beta N acetylglucosaminidase F2 | 71,647 | 2 | 3.0722 |
| A0A0A2VDW0 | Concanavalin A-like lectin/glucanases superfamily | 21,878 | 2 | 8.7379 |
| A0A0A2VFU8 | Thioredoxin reductase | 46,622 | 2 | 4.1475 |
| A0A0A2VG14 | Chitinase D | 34,889 | 2 | 8.7879 |
| A0A0A2VKM6 | Metallo-Zn-Carboxypeptidase A-like protein (M14A) | 43,858 | 2 | 5.7789 |
| A0A0A2VMQ3 | Fluoride ion transporter | 7335 | 2 | 25 |
| A0A0A2VUX9 | Putative dipeptidyl peptidase 5 | 109,325 | 2 | 2.0121 |
| A0A0A2VWX1 | Alpha N arabinofuranosidase | 53,574 | 2 | 4.3738 |
| A0A0A2VZ64 | Glucan 1,3 beta glucosidase | 83,915 | 2 | 3.0968 |
| A0A0A2W0E3 | Tripeptidyl peptidase sed2 | 59,491 | 2 | 4.6181 |
| A0A0A2W0S0 | Lipase 1 | 51,056 | 2 | 4.7228 |
| A0A0A2V532 | Chitinase, Glycosyl hydrolase 18 family (GH18) | 39,980 | 1 | 3.0137 |
| A0A0A2VAD7 | Small secreted protein | 14,925 | 1 | 7.8571 |
| A0A0A2VC64 | Glutathione reductase | 51,184 | 1 | 2.537 |
| A0A0A2VCD5 | Transcription factor Opi1 | 24,366 | 1 | 4.2735 |
| A0A0A2VCZ9 | Neutral cholesterol ester hydrolase | 36,385 | 1 | 2.1407 |
| A0A0A2VDJ2 | Hypoxia up-regulated protein 1 | 112,148 | 1 | 1.564 |
| A0A0A2VES3 | Regulation of enolase protein 1 (DUF1349) | 20,815 | 1 | 5.8201 |
| A0A0A2VFI0 | Cytochrome oxidase assembly protein 1 | 24,527 | 1 | 4.5872 |
| A0A0A2VFS0 | Dolichyl phosphate mannose protein mannosyltransferase 1 | 100,507 | 1 | 0.5543 |
| A0A0A2VGF4 | Putative serine protease S8 K12H47-like | 57,301 | 1 | 2.3438 |
| A0A0A2VJQ2 | Assimilatory nitrite reductase (NirD) small subunit | 8982 | 1 | 13.6364 |
| A0A0A2VM88 | Putative alpha beta glucosidase agdC | 54,004 | 1 | 2.459 |
| A0A0A2VMZ6 | RNA pyrophosphohydrolase (Nudix) | 18,709 | 1 | 8.7209 |
| A0A0A2VR04 | Putative 60S ribosomal protein MRP49 | 22,558 | 1 | 3.9216 |
| A0A0A2VRG8 | Putative U3 small nucleolar RNA-associated protein 13 | 166,473 | 1 | 0.3979 |
| A0A0A2VW39 | tRNA Guanine 37 N1 methyltransferase | 28,437 | 1 | 1.9608 |
| A0A0A2W0I5 | Pre rRNA processing protein esf1 | 84,406 | 1 | 1.7196 |
| A0A0A2W3A3 | Beta glucosidase | 94,734 | 1 | 1.6018 |
| A0A0A2W457 | Glycerol 3 phosphate dehydrogenase | 56,490 | 1 | 1.5936 |
| A0A0A2W4E7 | Ribose import ATP binding protein RbsA | 46,279 | 1 | 1.6667 |
| A0A0A2WIE5 | Aspartic protease | 37,508 | 1 | 2.5352 |
| Accession | Peptide Counts | ANOVA (p) | Description | Intact | Cuticle | Cuticle | Cuticle | Cuticle | Cuticle | Cuticle | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bb 813 | Stdev | Bb 813 | Stdev | Bb BG | Stdev | Bb 949 | Stdev | Bb 893 | Stdev | Bb 715 | Stdev | Bb 678 | Stdev | ||||
| A0A0A2VJD2 | 2 | 2.11E-15 | unknown protein | 508.94 | 48.82 | 81.64 | 3.57 | 57.58 | 1.36 | 42.54 | 1.64 | 34.23 | 3.94 | 28.18 | 3.66 | 18.11 | 2.98 |
| A0A0A2W0R3 | 23 | 0 | Pr1A Alkaline serine protease | 267.04 | 31.07 | 41.88 | 2.67 | 99.87 | 0.89 | 52.44 | 0.27 | 20.05 | 0.85 | 45 | 1.09 | 62.18 | 0.65 |
| A0A0A2VQS8 | 40 | 0 | Cell wall mannoprotein 1 | 241.81 | 3.24 | 739.44 | 12.22 | 115.12 | 2.38 | 217.32 | 3.23 | 80.16 | 0.48 | 45.72 | 2.44 | 45.13 | 1.08 |
| A0A0A2VHX5 | 8 | 1.11E-16 | Six-hairpin alpha-glycosidase GH13 | 167.49 | 9.82 | 33.39 | 0.39 | 64.16 | 2.01 | 47.65 | 1.94 | 26.81 | 0.48 | 28.57 | 1.82 | 44.26 | 0.95 |
| P00924 | 41 | 1.57E-13 | Enolase 1 (Saccharomyces cerevisiae) | 100 | 1.43 | 100 | 3.02 | 100 | 2.68 | 100 | 1.57 | 100 | 2.33 | 100 | 9.12 | 100 | 3.61 |
| A0A0A2VW01 | 15 | 1.96E-12 | Pr1B Alkaline protease 1 | 81.89 | 4.07 | 96.01 | 2.03 | 70.87 | 1.34 | 88.23 | 1.74 | 62.95 | 1.6 | 32.48 | 2.63 | 43.25 | 1.34 |
| A0A0A2VG14 | 10 | 0 | Chitinase D | 75.71 | 2.34 | 48.53 | 0.69 | 48.46 | 0.9 | 68.41 | 1.54 | 47.39 | 0.22 | 13.82 | 0.98 | 14.18 | 0.67 |
| A0A0A2VDM1 | 8 | 9.23E-10 | Putative glycosidase crf1 | 60.45 | 1.49 | 131.68 | 0.38 | 115.36 | 4.46 | 104.15 | 1.18 | 97.46 | 4.35 | 69.12 | 9.38 | 74.13 | 4.04 |
| A0A0A2VPJ3 | 17 | 8.69E-13 | Extracellular aldono-Lactonase YkgB | 48.86 | 2.38 | 36.56 | 0.94 | 98.42 | 4.14 | 100.11 | 0.49 | 57.65 | 0.36 | 46.46 | 3.42 | 43.88 | 1.93 |
| A0A0A2VRL8 | 17 | 1.68E-13 | l-ascorbate peroxidase | 43.14 | 0.62 | 95.94 | 3.58 | 198.51 | 3 | 180.37 | 3.08 | 71 | 0.82 | 49.6 | 5.06 | 65.99 | 1.46 |
| A0A0A2W9X8 | 5 | 8.81E-09 | Sialidase | 42.84 | 8.4 | 12.39 | 0.51 | 15.17 | 0.85 | 16.26 | 2.72 | 9.5 | 1.15 | 12.28 | 0.68 | 17.55 | 0.73 |
| A0A0A2VIV9 | 5 | 0 | Isochorismatase-like (Cysteine hydrolases) protein ycaC | 39.05 | 2.27 | 3.32 | 0.44 | 2.12 | 0.12 | 1.5 | 0.09 | 0.84 | 0.06 | 0.61 | 0.05 | 0.91 | 0.07 |
| A0A0A2VL34 | 2 | 0 | Putative glucan endo-1,3-beta-glucosidase eglC | 37.09 | 0.84 | 9.07 | 0.32 | 7.54 | 0.22 | 14.27 | 0.39 | 6.76 | 0.21 | 1.4 | 0.18 | 1.69 | 0.09 |
| A0A0A2VJ73 | 11 | 1.69E-13 | Alpha-galactosidase | 35.98 | 3.93 | 28.71 | 0.91 | 7 | 0.58 | 12.49 | 0.18 | 10.14 | 0.71 | 2.73 | 0.76 | 3.27 | 0.18 |
| A0A0A2VSY1 | 2 | 0 | Subtilisin-like proteinase Spm1 | 35.86 | 1.1 | 4.14 | 0.28 | 6.74 | 0.32 | 4.84 | 0.02 | 3.02 | 0.1 | 2.6 | 0.21 | 3.13 | 0.16 |
| A0A0A2VGF4 | 5 | 6.83E-10 | Putative serine protease S28 (K12H4.7) | 30.8 | 1.58 | 21.24 | 1.24 | 26.17 | 1.03 | 23.87 | 0.15 | 12.15 | 0.34 | 12.71 | 1.52 | 16.64 | 0.6 |
| A0A0A2W0S0 | 2 | 0 | Lipase 1 | 28.66 | 0.88 | 6.82 | 0.21 | 2.74 | 0.06 | 2.83 | 0.06 | 1.99 | 0.2 | 1.31 | 0.16 | 2.12 | 0.07 |
| A0A0A2V928 | 13 | 2.01E-12 | Catalase-peroxidase | 28.1 | 0.97 | 13.73 | 0.6 | 20.14 | 0.04 | 27.45 | 1.42 | 13.54 | 0.81 | 10.64 | 0.2 | 13.91 | 0.26 |
| A0A0A2VUK5 | 2 | 3.44E-15 | Endo-chitosanase | 27.53 | 0.56 | 20.96 | 1.29 | 2.81 | 0.1 | 3.18 | 0.18 | 1.84 | 0.24 | 1.08 | 0.26 | 2.24 | 0.36 |
| A0A0A2VL54 | 4 | 2.22E-16 | Autolysin (Lysine Motif, LysM protein) | 25.57 | 1.91 | 19.84 | 0.53 | 26.24 | 0.88 | 24.45 | 0.94 | 15.83 | 0.95 | 71.63 | 5.57 | 59.59 | 2.1 |
| A0A0A2V8V1 | 3 | 6.66E-16 | Vacuolar protease A | 25.04 | 0.54 | 10.77 | 0.61 | 5.58 | 0.25 | 7.29 | 0.01 | 4.8 | 0.23 | 2.74 | 0.26 | 4.18 | 0.19 |
| A0A0A2VC64 | 1 | 1.11E-15 | Glutathione reductase | 24.96 | 1.19 | 17.78 | 0.77 | 43.06 | 1.81 | 29.62 | 0.36 | 13.94 | 0.97 | 41.15 | 4.7 | 89.06 | 4.45 |
| A0A0A2VWW0 | 6 | 1.17E-13 | Cell_wall_mannoprotein_1 | 23.67 | 1.06 | 26.27 | 0.91 | 28.03 | 0.25 | 45.77 | 1.2 | 17.12 | 0.08 | 32.77 | 2.23 | 24.19 | 0.3 |
| A0A0A2V7C2 | 6 | 9.10E-15 | Beta-1,3 exoglucanase | 22.11 | 1.2 | 12.72 | 0.16 | 11.1 | 0.09 | 19.31 | 0.73 | 12.48 | 0.49 | 9.91 | 0.22 | 11.66 | 0.33 |
| A0A0A2VMQ3 | 1 | 0.02458 | Fluoride ion transporter | 19.72 | 1.12 | 8.4 | 3.16 | 3.8 | 0.98 | 7.24 | 3.82 | 4.53 | 3.42 | 6.5 | 2.18 | 6.17 | 1.31 |
| A0A0A2VJ18 | 10 | 7.17E-14 | Reticulocyte-binding protein 2 a | 18.09 | 0.33 | 63.6 | 1.29 | 74.32 | 1.83 | 65.55 | 4.05 | 63.73 | 5.35 | 52.53 | 1.55 | 58.97 | 1.71 |
| A0A0A2VAM6 | 1 | 2.81E-14 | Putative glucan endo-1,3-beta-glucosidase eglC | 16.03 | 0.59 | 7.76 | 0.33 | 9.88 | 0.24 | 8.71 | 0.11 | 2.39 | 0.48 | 2.82 | 0.17 | 1.54 | 0.06 |
| A0A0A2W3A3 | 5 | 5.38E-13 | Beta-glucosidase | 15.3 | 1.67 | 130.33 | 11.33 | 27.68 | 1.13 | 35.33 | 0.44 | 18.32 | 0.84 | 14.95 | 2.1 | 12.71 | 0.34 |
| A0A0A2VR76 | 4 | 2.29E-12 | Carboxypeptidase Y A | 13.82 | 1.27 | 9.87 | 0.29 | 27.52 | 1.49 | 17.53 | 0.29 | 8.67 | 0.48 | 20.36 | 1.58 | 10.61 | 0.34 |
| A0A0A2VFU8 | 5 | 9.04E-11 | Thioredoxin reductase | 12.18 | 0.89 | 7.9 | 0.16 | 13.3 | 0.23 | 7.96 | 0.69 | 3.68 | 0.23 | 7.42 | 1.02 | 10.84 | 0.29 |
| A0A0A2VWX1 | 4 | 5.55E-16 | Alpha-N-arabinofuranosidase | 12.01 | 0.51 | 14.49 | 0.3 | 72.11 | 1.86 | 38.57 | 0.78 | 19.18 | 0.15 | 11.18 | 0.93 | 16.19 | 0.2 |
| A0A0A2VZS6 | 2 | 2.69E-06 | Putative J domain-containing protein C3E7.11 c | 11.57 | 0.1 | 10 | 0.29 | 14.16 | 0.55 | 11.2 | 1.71 | 8.42 | 0.24 | 9.66 | 0.63 | 11.22 | 0.41 |
| A0A0A2VS40 | 2 | 4.04E-10 | Protein NIF3 (NGG1p interacting factor 3) | 10.84 | 3.32 | 3.86 | 0.27 | 5.43 | 0.35 | 15.87 | 0.48 | 5.62 | 0.37 | 1.94 | 0.34 | 0.98 | 0.3 |
| A0A0A2VNH3 | 3 | 2.92E-13 | Beta-galactosidase | 10.83 | 0.49 | 13.32 | 0.9 | 59.3 | 1.86 | 33.18 | 0.48 | 16.36 | 0.76 | 8.17 | 1.06 | 7.79 | 0.38 |
| A0A0A2VSI9 | 1 | 1.12E-12 | Putative Zn(II)2Cys6 transcription factor | 10.63 | 0.76 | 6.31 | 0.23 | 5.55 | 0.38 | 8.48 | 0.05 | 5.34 | 0.36 | 7.65 | 0.36 | 8.82 | 0.42 |
| A0A0A2VWS4 | 11 | 3.11E-15 | Alkaline phosphatase H | 10.34 | 0.45 | 21.51 | 0.14 | 41.28 | 1.21 | 22.39 | 0.69 | 20.16 | 0.36 | 34.08 | 4 | 36.46 | 1.44 |
| A0A0A2VAJ1 | 8 | 1.10E-09 | Tripeptidyl-peptidase sed1 | 9.78 | 0.66 | 9.15 | 0.45 | 12.12 | 0.5 | 6.77 | 0.1 | 5.5 | 0.29 | 4.92 | 0.47 | 5.14 | 0.22 |
| A0A0A2VMK7 | 2 | 1.92E-14 | AP-1-like transcription factor | 8.88 | 0.43 | 5.01 | 0.32 | 50.81 | 2.31 | 22.31 | 0.12 | 8.58 | 0.56 | 4.09 | 0.59 | 7.17 | 0.36 |
| A0A0A2VGH0 | 2 | 1.43E-08 | Glucan endo-1,3-beta-glucosidase | 7.38 | 0.82 | 7.2 | 0.35 | 7.87 | 0.32 | 7.2 | 0.15 | 5.65 | 0.58 | 8.21 | 1.52 | 7.21 | 0.64 |
| A0A0A2VJP2 | 2 | 1.31E-07 | ATP-dependent RNA helicase glh-2 | 7.14 | 1.24 | 11.55 | 0.36 | 8.05 | 0.23 | 9.7 | 0.18 | 8.44 | 0.3 | 6.39 | 0.73 | 6.22 | 0.22 |
| A0A0A2VZ64 | 2 | 3.61E-07 | Glucan 1,3-beta-glucosidase | 6.85 | 0.98 | 7.54 | 0.38 | 9.81 | 0.6 | 7.66 | 0.15 | 5.3 | 0.33 | 5.9 | 0.53 | 8.29 | 0.19 |
| A0A0A2VUJ3 | 3 | 0 | Glutamine-tRNA ligase | 6.45 | 0.17 | 6.36 | 0.24 | 11.56 | 0.48 | 22.08 | 0.27 | 10.71 | 0.38 | 48.56 | 5.78 | 44.18 | 1.51 |
| A0A0A2W0E3 | 5 | 1.69E-09 | Tripeptidyl-peptidase sed2 | 4.31 | 0.23 | 15.1 | 0.84 | 27.45 | 1.2 | 12 | 1.75 | 5.68 | 0.92 | 8.26 | 0.88 | 19.98 | 0.48 |
| A0A0A2VBW0 | 2 | 5.20E-05 | Translation initiation factor IF-2 | 3.39 | 0.2 | 18.95 | 3.7 | 61.44 | 13.35 | 31.76 | 10.98 | 11.81 | 6.98 | 17.17 | 1.78 | 35.49 | 1.88 |
| A0A0A2V6D8 | 4 | 3.54E-14 | Flagellar motor protein MotB (i.e., XP_007808661) | 2.82 | 0.45 | 6.27 | 0.38 | 35.33 | 1.72 | 13.78 | 0.1 | 8.35 | 0.81 | 25.02 | 3.04 | 13.94 | 0.84 |
| A0A0A2V5R3 | 2 | 0 | Glycosyltransferase family 90 protein | 2.2 | 0.17 | 6.55 | 0.22 | 3.82 | 0.25 | 37.63 | 0.58 | 40.78 | 2.24 | 4.29 | 0.33 | 3.43 | 0.11 |
| A0A0A2VPN1 | 1 | 9.31E-12 | BTB/POZ domain zinc finger transcription factor | 1.12 | 0.46 | 14.77 | 0.58 | 4.1 | 0.58 | 7.31 | 0.3 | 4.61 | 0.11 | 0.14 | 0.03 | 2.15 | 0.16 |
| A0A0A2W8Q3 | 1 | 8.40E-12 | Brefeldin A resistance protein | 1.06 | 0.35 | 4.07 | 0.36 | 2.99 | 0.16 | 7.92 | 0.24 | 3.08 | 0.5 | 9.9 | 1.32 | 8.06 | 0.66 |
| A0A0A2VIC5 | 1 | 4.44E-16 | Nickel/cobalt efflux system rcnA | 1.01 | 0.22 | 2.16 | 0.2 | 1.14 | 0.07 | 2.25 | 0.16 | 0.01 | 0.01 | 12.33 | 1.21 | 5.93 | 0.24 |
| A0A0A2W2R1 | 1 | 4.33E-15 | Glucan synthesis regulatory protein | 0.83 | 0.04 | 2.27 | 0.27 | 25.72 | 1.04 | 14.57 | 0.55 | 4.46 | 0.15 | 1.55 | 0.25 | 2.58 | 0.24 |
| A0A0A2VD91 | 2 | 1.27E-14 | alpha-1,2-Mannosidase | 0.01 | 0 | 1.86 | 0.12 | 1.21 | 0.03 | 1.43 | 0.17 | 0.06 | 0.02 | 7.07 | 1.57 | 6.32 | 1.35 |
| 0A0A2VEQ5 | 1 | 0 | DNA polymerase III subunits gamma/tau | 0 | 0 | 0.53 | 0.08 | 0.62 | 0.09 | 0.63 | 0.07 | 0.01 | 0.01 | 5.61 | 0.74 | 9.17 | 0.13 |
| A0A0A2VWT7 | 3 | 0 | Phospholipase A2 | 0.18 | 0.02 | 4.35 | 0.27 | 15.73 | 0.7 | 11.09 | 0.09 | 7.21 | 0.29 | 8.59 | 0.91 | 3.32 | 0.26 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dionisio, G.; Kryger, P.; Steenberg, T. Label-Free Differential Proteomics and Quantification of Exoenzymes from Isolates of the Entomopathogenic Fungus Beauveria bassiana. Insects 2016, 7, 54. https://doi.org/10.3390/insects7040054
Dionisio G, Kryger P, Steenberg T. Label-Free Differential Proteomics and Quantification of Exoenzymes from Isolates of the Entomopathogenic Fungus Beauveria bassiana. Insects. 2016; 7(4):54. https://doi.org/10.3390/insects7040054
Chicago/Turabian StyleDionisio, Giuseppe, Per Kryger, and Tove Steenberg. 2016. "Label-Free Differential Proteomics and Quantification of Exoenzymes from Isolates of the Entomopathogenic Fungus Beauveria bassiana" Insects 7, no. 4: 54. https://doi.org/10.3390/insects7040054