Genome Investigations of Vector Competence in Aedes aegypti to Inform Novel Arbovirus Disease Control Approaches
Abstract
:1. Introduction
2. Gene by Environment Interactions Determine Vector Competence
3. Quantitative Genetics of Vector Competence
3.1. Innate Immune Response Defines Vector Competence
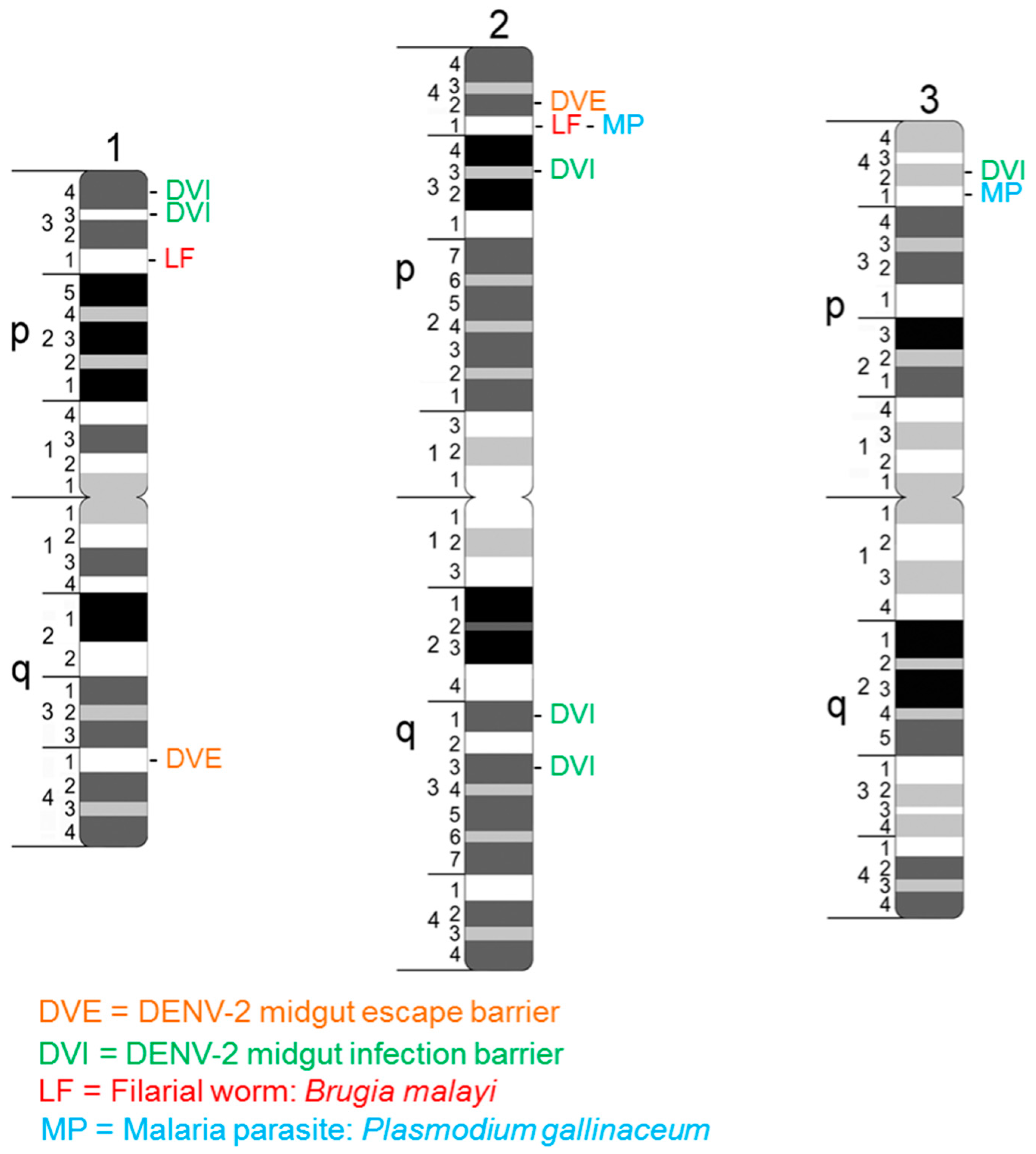
3.2. Primary Conditioners of Vector Competence Identified as Quantitative Trait Loci
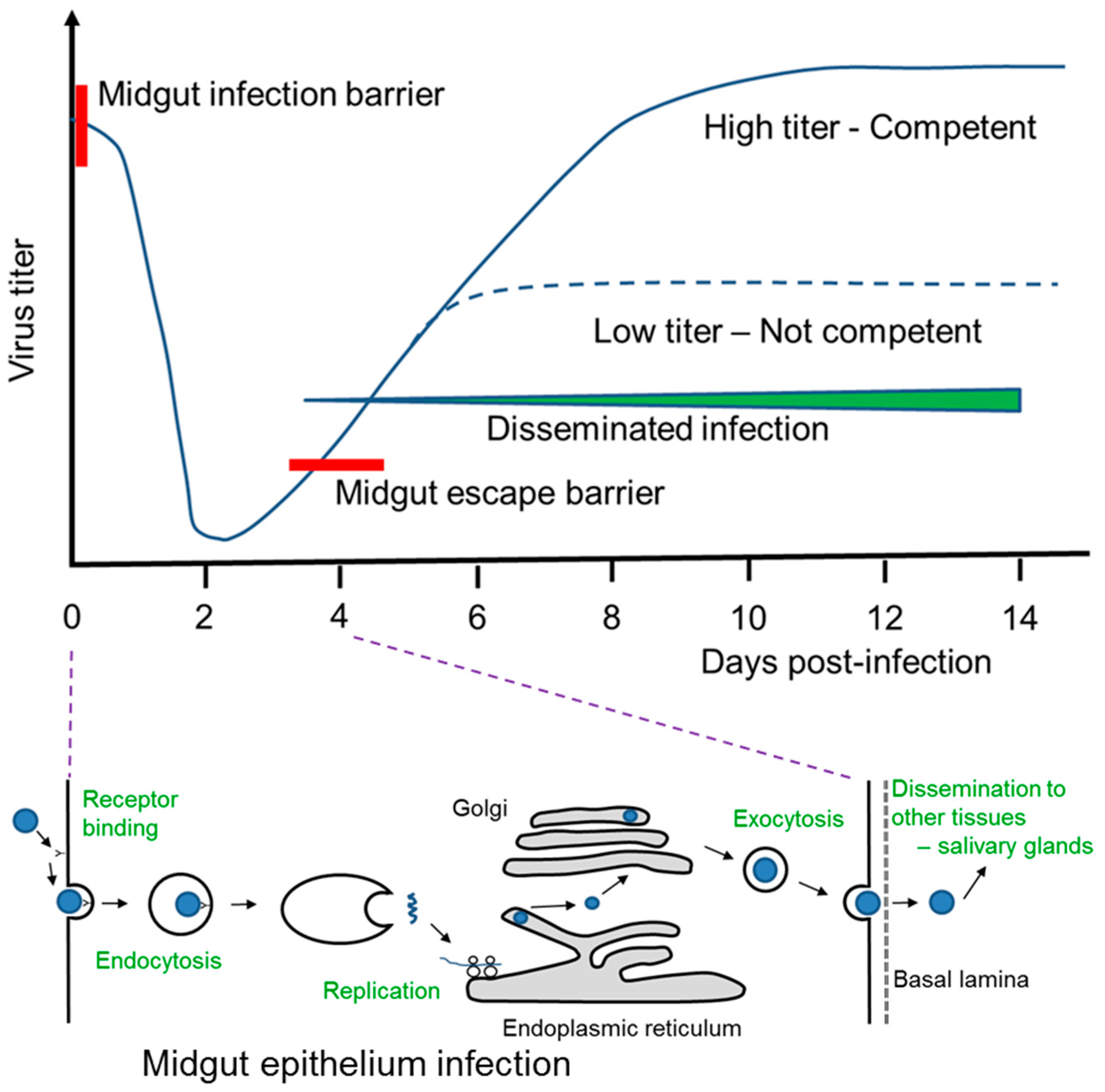
4. A. aegypti and DENV Interaction Post-Blood Feeding on Infected Human Host
5. Functional Genomics of Innate Immune Response to DENV
6. Genome Coevolution and Vector Competence
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DENV | Dengue virus |
| GM | Genotype of mosquito |
| GV | Genotype of virus |
| E | Environment |
| VC | Vectorial capacity |
| QTL | Quantitative trait locus |
| MIB | Midgut infection barrier |
| MEB | Midgut escape barrier |
| EIP | Extrinsic incubation period |
| RNAi | RNA interference |
| WNV | West Nile virus |
| YF | Yellow fever virus |
References
- Brady, O.B.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Vasconcelos, P.F.C. Yellow fever. J. Clin. Virol. 2015, 64, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Caglioti, C.; Lalle, E.; Castilletti, C.; Carletti, F.; Capobianchi, M.R.; Bordi, L. Chikungunya virus infections: An overview. New Microbiol. 2013, 36, 211–227. [Google Scholar] [PubMed]
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.-Y.; Vasilakis, N. Zika virus: History, emergence, biology, and prospects for control. Antivir. Res. 2016, 130, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Cheng, G. Vaccines and immunization strategies for dengue prevention. Emerg. Microbes Infect. 2016, 5, e77. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.J.; Rothman, A.L. Trials and tribulations on the path to developing a dengue vaccine. Vaccine 2015, 33, D24–D31. [Google Scholar] [CrossRef] [PubMed]
- Hemingway, J.; Ranson, H. Insecticide resistance in insect vectors of human disease. Annu. Rev. Entomol. 2000, 45, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Alphey, L.; McKemey, A.; Nimmo, D.; Oviedo, M.N.; Lacroix, R.; Matzen, K.; Beech, C. Genetic control of Aedes mosquitoes. Pathog. Glob. Health 2013, 107, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Alphey, L. Genetic control of mosquitoes. Annu. Rev. Entomol. 2014, 59, 205–224. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.E.; Clem, R.J.; Passarelli, A.L. Novel genetic and molecular tools for the investigation and control of dengue virus transmission by mosquitoes. Curr. Trop. Med. Rep. 2014, 1, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.F.; McKemy, A.R.; Nimmo, D.; Curtis, Z.; Black, I.; Morgan, S.A.; Oviedo, M.N.; Lacroix, R.; Naish, N.; Morrison, N.I.; et al. Successful suppression of a field mosquito population by sustained release of engineering male mosquitoes. Nat. Biotechnol. 2012, 30, 828–830. [Google Scholar] [CrossRef] [PubMed]
- Wise De Valdez, M.R.; Nimmo, D.; Betz, J.; Gong, H.-F.; James, A.A.; Alphey, L.; Black, W.C., IV. Genetic elimination of dengue vector mosquitoes. Proc. Natl. Acad. Sci. USA 2011, 108, 4772–4775. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 2011, 476, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Tabachnick, W.J. Nature, nurture and evolution of intra-species variation in mosquito arbovirus transmission competence. Int. J. Environ. Res. Public Health 2013, 10, 249–277. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.E.; Kantor, A.M.; Passarelli, A.L.; Clem, R.J. Tissue barriers to arbovirus infection in mosquitoes. Viruses 2015, 7, 3741–3767. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C., IV; Severson, D.W. Genetics of vector competence. In Biology of Disease Vectors, 2nd ed.; Marquardt, W.C., Ed.; Elsevier: London, UK, 2005; pp. 415–448. [Google Scholar]
- Lambrechts, L.; Chevillon, C.; Albright, R.G.; Thaisomboonsuk, B.; Richardson, J.H.; Jarman, R.G.; Scott, T.W. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol. Biol. 2009. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, L. Quantitative genetics of Aedes aegypti vector competence for dengue viruses: Towards a new paradigm? Trends Parasitol. 2011, 27, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Lequime, S.; Fontaine, A.; Gouilh, M.A.; Moltini-Conclois, I.; Lambrechts, L. Genetic drift, purifying selection and vector genotype shape dengue virus intra-host genetic diversity in mosquitoes. PLoS Genet. 2016, 12, e1006111. [Google Scholar] [CrossRef] [PubMed]
- Bennett, K.E.; Olson, K.E.; de Lourdes Munoz, M.; Fernandez-Salas, I.; Farfan-Ale, J.A.; Higgs, S.; Black, W.C., IV; Beaty, B.J. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am. J. Trop. Med. Hyg. 2002, 67, 85–92. [Google Scholar] [PubMed]
- Diallo, M.; Sall, A.A.; Moncayo, A.C.; Ba, Y.; Fernandez, Z.; Ortiz, D.; Coffey, L.L.; Mathiot, C.; Tesh, R.B.; Weaver, S.C. Potential role of sylvatic and domestic African mosquito species in dengue emergence. Am. J. Trop. Med. Hyg. 2005, 73, 445–449. [Google Scholar] [PubMed]
- Failloux, A.-B.; Vazeille, M.; Rodhain, F. Geographic genetic variation in populations of the dengue virus vector Aedes aegypti. J. Mol. Evol. 2002, 55, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, C.M.; Melo, F.F.; Bezerra, M.T.; Chaves, B.A.; Silva, B.M.; Silva, L.D.; Pessanha, J.E.M.; Arias, J.R.; Secundino, N.F.C.; Norris, D.E.; et al. Distinct variation in vector competence among nine field populations of Aedes aegypti from a Brazilian dengue-endemic risk city. Parasites Vectors 2014. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J.; Nalim, S.; Tan, R.; Saipan, H.; Saroso, J.S. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am. J. Trop. Med. Hyg. 1979, 28, 1045–1052. [Google Scholar] [PubMed]
- Schneider, J.R.; Mori, A.; Romero-Severson, J.; Chadee, D.D.; Severson, D.W. Investigations of dengue-2 susceptibility and body size among Aedes aegypti populations. Med. Vet. Entomol. 2007, 21, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Sumanochitrapon, W.; Strickman, D.; Sithiprasasna, R.; Kittayapong, P.; Innis, B.L. Effect of size and geographic origin of Aedes aegypti on oral infection with dengue-2 virus. Am. J. Trop. Med. Hyg. 1998, 58, 283–286. [Google Scholar] [PubMed]
- Tardieux, I.; Poupel, O.; Lapchin, L.; Rodhain, F. Variation among strains of Aedes aegypti in susceptibility to oral infection with denge virus type 2. Am. J. Trop. Med. Hyg. 1990, 43, 308–313. [Google Scholar] [PubMed]
- Anderson, J.R.; Rico-Hesse, R. Aedes aegypti vectorial capacity is determined by the infecting genotype of the dengue virus. Am. J. Trop. Med. Hyg. 2006, 75, 886–892. [Google Scholar] [PubMed]
- Armstrong, P.M.; Rico-Hesse, R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am. J. Trop. Med. Hyg. 2003, 68, 539–544. [Google Scholar] [PubMed]
- Fernandes da Moura, A.J.; Varjal de Melo Santos, M.A.; Oliveira, C.M.F.; Guedes, D.R.D.G.; de Carvalho-Leandro, D.; da Cruz Brito, M.L.; Rocha, H.D.R.; Gomez, L.F.; Ayres, C.F.J. Vector competence of the Aedes aegypti population from Santiago Island, Cape Verde, to different serotypes of dengue virus. Parasites Vectors 2015. [Google Scholar] [CrossRef]
- Gaye, A.; Faye, O.; Diagne, C.T.; Faye, O.; Diallo, D.; Weaver, S.C.; Sall, A.A.; Diallo, M. Oral susceptibility of Aedes aegypti (Diptera: Culicidae) from Senegal for dengue serotypes 1 and 3 viruses. Trop. Med. Int. Health 2014, 19, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Khoo, C.C.H.; Doty, J.B.; Held, N.L.; Olson, K.E.; Franz, A.W.E. Isolation of midgut escape mutants of two American genotype dengue 2 viruses from Aedes aegypti. Virol. J. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, L.; Fansiri, T.; Pongsiri, A.; Thalsomboonsuk, B.; Klungthong, C.; Richardson, J.H.; Ponlawat, A.; Jarman, R.G.; Scott, T.W. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J. Virol. 2012, 86, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Carrington, L.B.; Seifert, S.N.; Armijos, M.V.; Lambrechts, L.; Scott, T.W. Reduction of Aedes aegypti vector competence for dengue virus under large temperature fluctuations. Am. J. Trop. Med. Hyg. 2013, 88, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Paupy, C.; Chantha, N.; Vazeille, M.; Reynes, J.-M.; Rodhain, F.; Failloux, A.-B. Variation over space and time of Aedes aegypti in Phnom Penh (Cambodia): Genetic structure and oral susceptibility to a dengue virus. Genet. Res. Camb. 2003, 82, 171–182. [Google Scholar] [CrossRef]
- Vazeille, M.; Gaborit, P.; Mousson, L.; Girod, R.; Failloux, A.-B. Competitive advantage of a dengue 4 virus when co-infecting the mosquito Aedes aegypti with dengue 1 virus. BMC Infect. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C., IV; Moore, C.G. Population biology as a tool to study vector-borne diseases. In Biology of Disease Vectors, 2nd ed.; Marquardt, W.C., Ed.; Elsevier: London, UK, 2005; pp. 187–206. [Google Scholar]
- Smith, D.L.; Battle, K.E.; Hay, S.I.; Barker, C.M.; Scott, T.W.; McKensie, F.E. Ross, Macdonald, and a theory for the dynamics and control of mosquito-transmitted pathogens. PLoS Pathog. 2012, 8, e1002588. [Google Scholar] [CrossRef] [PubMed]
- Tabachnick, W.J. Ecological effects on arbovirus-mosquito cycles of transmission. Curr. Opin. Virol. 2016, 21, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Maciel-de-Freitas, R.; Codeco, C.T.; Lourenco-de-Oliveira, R. Body size-associated survival and dispersal rates of Aedes aegypti in Rio de Janeiro. Med. Vet. Entomol. 2007, 21, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Nasci, R.S. The size of emerging and host-seeking Aedes aegypti and the relationship of size to blood-feeding success in the field. J. Am. Mosq. Control Assoc. 1986, 2, 61–62. [Google Scholar] [PubMed]
- Nasci, R.S. Influence of larval and adult nutrition on biting persistence in Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 1991, 31, 522–526. [Google Scholar] [CrossRef]
- Yan, G.; Severson, D.W.; Christensen, B.M. Costs and benefits of mosquito refractoriness to malaria parasites: Implications for genetic variability of mosquitoes and genetic control of malaria. Evolution 1997, 51, 441–450. [Google Scholar] [CrossRef]
- Coon, K.L.; Vogel, K.J.; Brown, M.R.; Strand, M.R. Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 2014, 23, 2727–2739. [Google Scholar] [CrossRef] [PubMed]
- Coon, K.L.; Brown, M.R.; Strand, M.R. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae). Parasit. Vectors 2016. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.W.; Amerasinghe, P.H.; Morrison, A.C.; Lorenz, L.H.; Clark, G.G.; Strickman, D.; Kittaypong, P.; Edman, J.D. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: Blood feeding frequency. J. Med. Entomol. 2000, 37, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.R.; Chadee, D.D.; Mori, A.; Romero-Severson, J.; Severson, D.W. Heritability and adaptive phenotypic plasticity of adult body size in the mosquito Aedes aegypti with implications for dengue vector competence. Infect. Genet. Evol. 2011, 11, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Barrera, B.; Amador, M.; Clark, G.C. Ecological factors influencing Aedes aegypti (Diptera: Culicidae) productivity in artificial containers in Salinas, Puerto Rico. J. Med. Entomol. 2006, 43, 484–492. [Google Scholar] [CrossRef]
- Hemme, R.R.; Tank, J.L.; Chadee, D.D.; Severson, D.W. Environmental conditions in water storage drums and influences on Aedes aegypti in Trinidad, West Indies. Acta Trop. 2009, 112, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Matsuse, I.T.; Takahashi, H.; Komukai, J.; Fukuda, T.; Suzuki, K.; Aratani, M.; Shira, Y.; Mogi, M. Effect of temperature on the development of Aedes aegypti and Aedes albopictus. Med. Entomol. Zool. 2002, 53, 53–58. [Google Scholar] [CrossRef]
- Keirans, J.E.; Fay, R.W. Effect of food and temperature on Aedes aegypti (L.) and Aedes triseriatus (SAY) larval development. Mosq. News 1968, 28, 338–341. [Google Scholar]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Govind, S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008, 15, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]
- Steinert, S.; Levashina, E.A. Intracellular immune responses of dipteran insects. Immunol. Rev. 2011, 240, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Tzou, P.; De Gregorio, E.; Lemaitre, B. How Drosophila combats microbial infection: A model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 2002, 5, 102–110. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.D.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef] [PubMed]
- Arjona, A.; Wang, P.; Montgomery, R.R.; Fikrig, E. Innate immune control of West Nile virus infection. Cell. Microbiol. 2011, 13, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Cirimotich, C.M.; Dong, Y.; Garver, L.S.; Sim, S.; Dimopoulos, G. Mosquito immune defenses against Plasmodium infection. Dev. Comp. Immunol. 2010, 34, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, G.; Muller, H.-M.; Levashina, E.A.; Kafatos, F.C. Innate immune defense against malaria infection in the mosquito. Curr. Opin. Immunol. 2001, 13, 79–88. [Google Scholar] [CrossRef]
- Fragkoudis, R.; Attarzadeh-Yazdi, G.; Nash, A.A.; Fazakerley, J.K.; Kohl, A. Advances in dissecting mosquito innate immune responses to arbovirus infection. J. Gen. Virol. 2009, 90, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.N.; Brackney, D.E.; Ebel, G.D. The role of innate immunity in conditioning mosquito susceptibility to West Nile Virus. Viruses 2013, 5, 3142–3170. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Jupatanakul, N.; Dimopoulos, G. Mosquito immunity against arboviruses. Viruses 2014, 6, 4479–4504. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C., IV; Bennett, K.E.; Gorrochotegui-Escalante, N.; Barillas-Mury, C.V.; Fernandez-Salas, I.; de Lourdes Munoz, M.; Farfan, J.A.; Olson, K.E.; Beaty, B.J. Flavivirus susceptibility in Aedes aegypti. Arch. Med. Res. 2002, 33, 379–388. [Google Scholar] [CrossRef]
- Severson, D.W.; Brown, S.E.; Knudson, D.L. Genetic and physical mapping in mosquitoes: Molecular approaches. Ann. Rev. Entomol. 2001, 46, 183–219. [Google Scholar] [CrossRef] [PubMed]
- Bosio, C.F.; Beaty, B.J.; Black, W.C., IV. Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am. J. Trop. Med. Hyg. 1998, 59, 965–970. [Google Scholar] [PubMed]
- Bennett, K.E.; Flick, D.; Fleming, K.H.; Jochim, R.; Beaty, B.J.; Black, W.C., IV. Quantitative trait loci that control dengue-2 virus dissemination in the mosquito Aedes aegypti. Genetics 2005, 170, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Bosio, C.F.; Fulton, R.E.; Salasek, M.L.; Beaty, B.J.; Black, W.C., IV. Quantative trait loci that control vector competence for dengue-2 virus in the mosquito Aedes aegypti. Genetics 2000, 156, 687–698. [Google Scholar] [PubMed]
- Gomez-Machorro, C.; Bennett, K.E.; del Lourdes Munoz, M.; Black, W.C., IV. Quantitative trait loci affecting dengue midgut infection barriers in an advanced intercross line of Aedes aegypti. Insect Mol. Biol. 2004, 13, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Beerntsen, B.T.; Severson, D.W.; Klinkhammer, J.A.; Kassner, V.A.; Christensen, B.M. Aedes aegypti: A quantitative trait locus (QTL) influencing filarial worm intensity is linked to QTL for susceptibility to other mosquito-borne pathogens. Exp. Parasitol. 1995, 81, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Severson, D.W.; Mori, A.; Zhang, Y.; Christensen, B.M. Chromosomal mapping of two loci affecting filarial worm susceptibility in Aedes aegypti. Insect Mol. Biol. 1994, 3, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Severson, D.W.; Thathy, V.; Mori, A.; Zhang, Y.; Christensen, B.M. Restriction fragment length polymorphism mapping of quantitative trait loci for malaria parasite susceptibility in the mosquito Aedes aegypti. Genetics 1995, 139, 1711–1717. [Google Scholar] [PubMed]
- Zhong, D.; Menge, D.M.; Temu, E.A.; Chen, H.; Yan, G. Amplified fragment length polymorphism mapping of quantitative trait loci for malaria parasite susceptibility in the yellow fever mosquito Aedes aegypti. Genetics 2006, 173, 1337–1345. [Google Scholar] [CrossRef] [PubMed]
- Timoshevskiy, V.A.; Severson, D.W.; deBruyn, B.S.; Black, W.C.; Sharakhov, I.V.; Sharakhova, M.V. An integrated linkage, chromosome, and genome map for the yellow fever mosquito Aedes aegypti. PLoS Negl. Trop. Dis. 2013, 7, e2052. [Google Scholar] [CrossRef] [PubMed]
- Wisser, R.J.; Balint-Kurti, P.J.; Nelson, R.J. The genetic architecture of disease resistance in maize: A synthesis of published studies. Phytopathology 2006, 96, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Rodenhuis-Zybert, I.A.; Wilschut, J.; Smit, J.M. Dengue virus life cycle: Viral and host factors modulating infectivity. Cell. Mol. Life Sci. 2010, 67, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. An update on mosquito cell expressed dengue virus receptor proteins. Insect Mol. Biol. 2012, 21, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Acosta, E.G.; Castilla, V.; Damonte, E.B. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J. Gen. Virol. 2008, 89, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Mosso, C.; Galvan-Mendoza, I.J.; Ludert, J.E.; del Angel, R.M. Endocytic pathway followed by dengue virus to infect the mosquito cell line C6/36 HT. Virology 2008, 378, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.; Molina-Cruz, A.; Salazar, M.I.; Black, W.B., IV. Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes aegypti. Am. J. Trop. Med. Hyg. 2006, 74, 132–141. [Google Scholar] [PubMed]
- Salazar, M.I.; Richardson, J.H.; Sanchez-Vargas, I.; Olson, K.E.; Beaty, B.J. Dengue virus type 2: Replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.E.; Wu, W.-K.; Verleye, D.; Rai, K.S. Midgut basal lamina thickness and dengue-1 virus dissemination rates in laboratory strains of Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 1993, 30, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Nguyet, M.N.; Kien, D.T.H.; Tuan, T.V.; Quyen, N.T.H.; Tran, C.N.B.; Thi, L.V.; Thi, D.L.; Nguyen, H.L.; Nguyen, H.T.C.; Nguyen, L.T.H.; et al. Host and viral features of human dengue cases shape the population of infected and infectious Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. USA 2013, 110, 9072–9077. [Google Scholar] [PubMed]
- Ye, Y.H.; Ng, T.S.; Frentiu, F.D.; Walker, T.; van den Hurk, A.F.; O’Neill, S.L.; Beebe, N.W.; McGraw, E.A. Comparative susceptibility of mosquito populations in North Queensland, Australia to oral infection with dengue virus. Am. J. Trop. Med. Hyg. 2014, 90, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.H.; Chenoweth, S.F.; Carrasco, A.M.; Allen, S.L.; Frentiu, F.D.; van den Hurk, A.F.; Beebe, N.W.; McGraw, E.A. Evolutionary potential of the extrinsic incubation period of dengue virus in Aedes aegypti. Evolution 2016. [Google Scholar] [CrossRef] [PubMed]
- Nene, V.; Wortman, J.R.; Lawson, D.; Haas, B.; Kodira, C.; Tu, Z.; Loftus, B.; Xi, Z.; Megy, K.; Grabherr, M.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 316, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef] [PubMed]
- Souza-Neto, J.A.; Sim, S.; Dimopoulos, G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 2009, 106, 17841–17846. [Google Scholar] [CrossRef] [PubMed]
- Colpitts, T.M.; Cox, J.; Vanlandingham, D.L.; Feitosa, F.M.; Cheng, G.; Kurscheid, S.; Wang, P.; Kishnan, M.N.; Higgs, S.; Fikrig, E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011, 7, e1002189. [Google Scholar] [CrossRef] [PubMed]
- Bonizzoni, M.; Dunn, W.A.; Campbell, C.L.; Olson, K.E.; Marinotti, O.; James, A.A. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS ONE 2012, 7, e50512. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensary apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef] [PubMed]
- Baron, O.L.; Ursic-Bedoya, R.J.; Lowenberger, C.A.; Ocampo, C.B. Differential gene expression from midguts of refractory and susceptible lines of the mosquito, Aedes aegypti, infected with dengue-2 virus. J. Insect Sci. 2008. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Gomez-Machorro, C.; Harker, B.W.; deBruyn, B.; Lovin, D.D.; Hemme, R.R.; Mori, A.; Romero-Severson, J.; Severson, D.W. Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection. PLoS Negl. Trop. Dis. 2011, 5, e1385. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Gomez-Machorro, C.; deBruyn, B.; Lovin, D.D.; Harker, B.W.; Romero-Severson, J.; Mori, A.; Severson, D.W. Influence of mosquito genotype on transcriptional response to dengue virus infection. Funct. Integr. Genom. 2014, 14, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, C.; Behura, S.K.; deBruyn, B.; Lovin, D.D.; Harker, B.W.; Gomez-Machorro, C.; Mori, A.; Romero-Severson, J.; Severson, D.W. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. PLoS ONE 2012, 7, e47350. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Jupatanakul, N.; Ramirez, J.L.; Kang, S.; Romero-Vivas, C.M.; Mohammed, H.; Dimopoulos, G. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl. Trop. Dis. 2013, 7, e2295. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Liu, Y.; Wang, P.; Xiao, X. Mosquito defense strategies against viral infection. Trends Parasitol. 2016, 32, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Clem, R.J. Arboviruses and apoptosis: The role of cell death in determining vector competence. J. Gen. Virol. 2016, 97, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.E.; Blair, C.D. Arbovirus-mosquito interactions: RNAi pathway. Curr. Opin. Virol. 2015, 15, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N.A. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 2008, 105, 19938–19943. [Google Scholar] [CrossRef] [PubMed]
- Green, A.M.; Beatty, P.R.; Hadjilaou, A.; Harris, E. Innate immunity to dengue virus infection and subversion of antiviral responses. J. Mol. Biol. 2014, 426, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Garcia, M.; Conde, R.; Bello-Bedoy, R.; Lanz-Mendoza, H. The damage threshold hypothesis and the immune strategies of insects. Infect. Genet. Evol. 2014, 24, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ayres, J.S.; Schneider, D.S. Tolerance of infections. Annu. Rev. Immunol. 2012, 30, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Eng, M.W.; van Zuylen, M.N.; Severson, D.W. Apoptosis-related genes control autophagy and influence DENV-2 infection in the mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2016, 76, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Jain, B.; Chaturvedi, U.C.; Jain, A. Role of intracellular events in the pathogenesis of dengue: An overview. Microb. Pathog. 2014, 69–70, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Lobo, F.P.; Mota, B.E.F.; Pena, S.D.J.; Azevedo, V.; Macedo, A.M.; Tauch, A.; Machado, C.R.; Franco, G.R. Virus-host coevolution: Common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE 2009, 4, e6282. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.T.; Carthew, R.W. A call to arms: Coevolution of animal viruses and host innate immune responses. Trends Genet. 2007, 23, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.T.; Imler, J.-L. The diversity of insect antiviral immunity: Insights from viruses. Curr. Opin. Microbiol. 2016, 32, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Intrinsic features of Aedes aegypti genes affect transcriptional responsiveness of mosquito genes to dengue virus infection. Infect. Genet. Evol. 2012, 12, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Calvo, E.; Pham, V.M.; Marinotti, O.; Andersen, J.F.; Ribeiro, J.M. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genom. 2009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, W.H. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol. Biol. Evol. 2004, 21, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.E.; Monie, T.P. Mice, men and the relatives: Cross-species studies underpin innate immunity. Open Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Dudas, G. The genetics of host-virus coevolution in invertebrates. Curr. Opin. Virol. 2014, 8, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Fransiri, T.; Pongsiri, A.; Klungthoung, C.; Ponlawat, A.; Thaisomboonsuk, B.; Jarman, R.G.; Scott, T.W.; Lambrechts, L. No evidence for local adaptation of dengue viruses to mosquito vector populations in Thailand. Evol. Appl. 2016, 9, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Sarro, J.; Li, P.; Mysore, K.; Severson, D.W.; Emrich, S.J.; Duman-Scheel, M. High-throughput cis-regulatory element discovery in the vector mosquito Aedes aegypti. BMC Genom. 2016. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Nucleotide substitutions in dengue virus serotypes from Asian and American countries: Insights into intracodon recombination and purifying selection. BMC Microbiol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lara-Ramirez, E.E.; Salazar, M.I.; Lopez-Lopez, M.J.; Salas-Benito, J.S.; Sanchez-Varela, A.; Guo, X. Large-scale genomic analysis of codon usage in dengue virus and evaluation of its phylogenetic dependence. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K.; Severson, D.W. Bicluster pattern of codon context usages between flavivirus and vector mosquito Aedes aegypti: Relevance to infection and transcriptome response of mosquito genes. Mol. Genet. Genom. 2014, 289, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.H.; Stauft, C.B.; Gorbatsevych, O.; Song, Y.; Ward, C.B.; Yurovsky, A.; Mueller, S.; Futcher, B.; Wimmer, E. Large-scale recoding of an arbovirus genome to rebalance its insect versus mammalian preference. Proc. Natl. Acad. Sci. USA 2015, 112, 4749–4754. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Deardorff, E.R.; Kenney, J.L.; Rossi, S.L.; Hanley, K.A.; Weaver, S.C. Mosquitoes put the brake on arbovirus evolution: Experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009. [Google Scholar] [CrossRef] [PubMed]
- Adelman, Z.N.; Jasinskiene, N.; James, A.A. Development and applications of transgenesis in the yellow fever mosquito, Aedes aegypti. Mol. Biochem. Parasitol. 2002, 121, 1–10. [Google Scholar] [CrossRef]
- Burt, B. Heritable strategies for controlling insect vectors of disease. Philos. Trans. R. Soc. B 2014. [Google Scholar] [CrossRef]
- Moreira, L.A.; Ghosh, A.K.; Abraham, E.G.; Jacobs-Lorena, M. Genetic transformation of mosquitoes: A quest for malaria control. Int. J. Parasitol. 2002, 32, 1599–1605. [Google Scholar] [CrossRef]
- Robinson, A.S.; Franz, G.; Atkinson, P.W. Insect transgenesis and its potential role in agriculture and human health. Insect Biochem. Mol. Biol. 2004, 34, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Dinglasan, R.R.; Jacobs-Lorena, M. Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 2008, 24, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Marinotti, O.; Sieglaff, D.; James, A.A. Molecular genetic manipulation of vector mosquitoes. Cell Host Microbe 2014, 4, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.W.; Takken, W.; Knols, B.G.J.; Boete, C. The ecology of genetically modified mosquitoes. Science 2002, 298, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Murdock, C.C.; Paaijmans, K.P.; Cox-Foster, D.; Read, A.F.; Thomas, M.B. Rethinking vector immunology: The role of environmental temperature in shaping resistance. Nat. Rev. Microbiol. 2012, 10, 869–876. [Google Scholar] [CrossRef] [PubMed]
| Study | Aedes aegypti strain(s) | Strain Susceptibility | Dengue Strain 3 | Mosquito Infection | Sample Point(s) Post-Infection and Tissues | Transcriptome Assay |
|---|---|---|---|---|---|---|
| [90] | Rockefeller | Susceptible 1 | A | oral | 10 days, midgut | Agilent microarrays |
| 10 days, carcass | ||||||
| [91] | Rockefeller | Susceptible | A | oral | 3 days, 7 days, whole body | Agilent microarrays |
| [95] | Cali | Susceptible | A | oral | 48 h, midgut | Suppressive subtractive hybridization |
| Cali | Refractory | |||||
| [92] | Rockefeller | Susceptible | A | injection | 1 day, 2 days, 7 days, whole body | NimbleGen |
| [96] | Moyo-R | Refractory | B | oral | 3 h, 18 h, whole body | NimbleGen |
| Moyo-S | ||||||
| [98] | Moyo-D | Refractory | B | oral | 1 h, 4 h, 1 day, 2 days, 4 days, midgut | Custom cDNA microarrays |
| Moyo-S | ||||||
| [93] | Chetumal | Susceptible | B | oral | 1 day, 4 days, midgut | Illumina, RNA-Seq |
| 14 days, salivary glands | ||||||
| 1 day, 4 days, 14 days, carcass | ||||||
| [94] | Rockefeller | Susceptible | A | oral | 14 days, salivary glands | Agilent microarrays |
| 14 days, carcass | ||||||
| [99] | Rockefeller | High 2 | A | oral | 7 days, midgut | Agilent microarrays |
| Orlando | Low | 7 days, carcass | ||||
| Waco | Low | |||||
| Puerto Rico | Intermediate | |||||
| Saint Kitts | Intermediate | |||||
| Por Fin | Intermediate | |||||
| Puerto Triunfo | High | |||||
| Singapore | High | |||||
| Bangkok | Low | |||||
| [97] | D2S3 | Susceptible | B | oral | 3 h, 3 days, midgut | Custom cDNA microarrays |
| Moyo-D | Refractory |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Severson, D.W.; Behura, S.K. Genome Investigations of Vector Competence in Aedes aegypti to Inform Novel Arbovirus Disease Control Approaches. Insects 2016, 7, 58. https://doi.org/10.3390/insects7040058
Severson DW, Behura SK. Genome Investigations of Vector Competence in Aedes aegypti to Inform Novel Arbovirus Disease Control Approaches. Insects. 2016; 7(4):58. https://doi.org/10.3390/insects7040058
Chicago/Turabian StyleSeverson, David W., and Susanta K. Behura. 2016. "Genome Investigations of Vector Competence in Aedes aegypti to Inform Novel Arbovirus Disease Control Approaches" Insects 7, no. 4: 58. https://doi.org/10.3390/insects7040058