Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi)
Abstract
1. Introduction
2. Results
2.1. Genome of ALPV
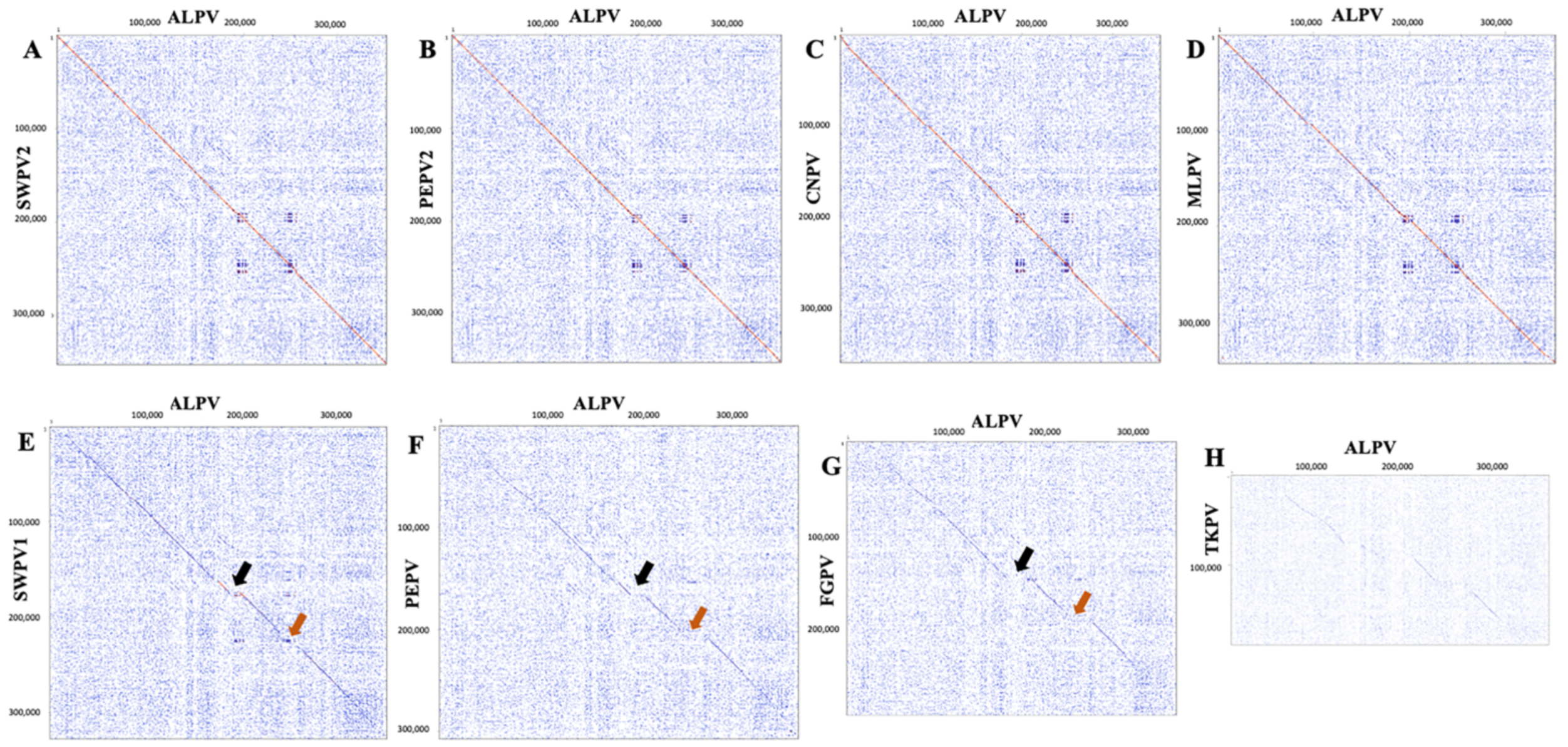
2.2. Genome Annotation and Comparative Analyses of ALPV
2.3. Evolutionary Relationships of ALPV
3. Discussion
4. Materials and Methods
4.1. Sampling and Virus Isolation
4.2. DNA Extraction and Sequencing
4.3. Genome Assembly and Annotation
4.4. Comparative Genomics
4.5. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Croxall, J.P.; Butchart, S.H.M.; Lascelles, B.E.N.; Stattersfield, A.J.; Sullivan, B.E.N.; Symes, A.; Taylor, P. Seabird conservation status, threats and priority actions: A global assessment. Bird Conserv. Int. 2012, 22, 1–34. [Google Scholar] [CrossRef]
- Croxall, J.P.; Gales, R. An assessment of the conservation status of albatrosses. In Albatross Biology and Conservation; Robertson, G., Gales, R., Eds.; Surrey Beatty & Sons: Chipping Norton, UK, 1998; pp. 46–65. [Google Scholar]
- Cooper, J.; Baker, G.B.; Double, M.C.; Gales, R.; Papworth, W.; Tasker, M.L.; Waugh, S.M. The agreement on the conservation of albatrosses and petrels: Rationale, history, progress and the way forward. Mar. Ornithol. 2006, 34, 1–5. [Google Scholar]
- Marcela, M.; Uhart, L.G.; Flavio, Q. Review of diseases (pathogen isolation, direct recovery and antibodies) in albatrosses and large petrels worldwide. Bird Conserv. Int. 2017, 28, 1–28. [Google Scholar] [CrossRef]
- Paleczny, M.; Hammill, E.; Karpouzi, V.; Pauly, D. Population Trend of the World’s Monitored Seabirds, 1950–2010. PLoS ONE 2015, 10, e0129342. [Google Scholar] [CrossRef]
- Phillips, R.A.; Gales, R.; Baker, G.B.; Double, M.C.; Favero, M.; Quintana, F.; Tasker, M.L.; Weimerskirch, H.; Uhart, M.; Wolfaardt, A. The conservation status and priorities for albatrosses and large petrels. Biol. Conserv. 2016, 201, 169–183. [Google Scholar] [CrossRef]
- BirdLife International. Diomedea Sanfordi. The IUCN Red List of Threatened Species 2018: E.T22728323A132656392. 2018. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T22728323A132656392.en (accessed on 30 January 2021).
- Sugishita, J. Northern royal albatross. In New Zealand Birds Online; Miskelly, C.M., Ed.; 2013; Available online: http://www.nzbirdsonline.org.nz/species/northern-royal-albatross (accessed on 20 January 2021).
- Halpern, B.S.; Walbridge, S.; Selkoe, K.A.; Kappel, C.V.; Micheli, F.; D’Agrosa, C.; Bruno, J.F.; Casey, K.S.; Ebert, C.; Fox, H.E.; et al. A global map of human impact on marine ecosystems. Science 2008, 319, 948–952. [Google Scholar] [CrossRef]
- Tuck, G.N.; Polacheck, T.; Croxall, J.P.; Weimerskirch, H. Modelling the impact of fishery by-catches on albatross populations. J. Appl. Ecol. 2001, 38, 1182–1196. [Google Scholar] [CrossRef]
- Baker, G.B.; Gales, R.; Hamilton, S.; Wilkinson, V. Albatrosses and petrels in Australia: A review of their conservation and management. Emu Austral Ornithol. 2002, 102, 71–97. [Google Scholar] [CrossRef]
- Lewison, R.L.; Crowder, L.B.; Read, A.J.; Freeman, S.A. Understanding impacts of fisheries bycatch on marine megafauna. Trends Ecol. Evol. 2004, 19, 598–604. [Google Scholar] [CrossRef]
- Rolland, V.; Barbraud, C.; Weimerskirch, H. Assessing the impact of fisheries, climate and disease on the dynamics of the Indian yellow-nosed Albatross. Biol. Conserv. 2009, 142, 1084–1095. [Google Scholar] [CrossRef]
- Department of Sustainability, Environment, Water, Population and Communities (2011) Background Paper, Population Status and Threats to Albatrosses and Giant Petrels Listed as Threatened under the Environment Protection and Biodiversity Conservation Act 1999. Commonwealth of Australia, Hobart. 2011. Available online: https://www.environment.gov.au/resource/background-paper-population-status-and-threats-albatrosses-and-giant-petrels-listed. (accessed on 29 January 2021).
- Illera, J.C.; Emerson, B.C.; Richardson, D.S. Genetic characterization, distribution and prevalence of avian pox and avian malaria in the Berthelot’s pipit (Anthus berthelotii) in Macaronesia. Parasitol. Res. 2008, 103, 1435–1443. [Google Scholar] [CrossRef]
- Lecis, R.; Secci, F.; Antuofermo, E.; Nuvoli, S.; Scagliarini, A.; Pittau, M.; Alberti, A. Multiple gene typing and phylogeny of avipoxvirus associated with cutaneous lesions in a stone curlew. Vet. Res. Commun. 2017, 1–7. [Google Scholar] [CrossRef]
- Woolaver, L.G.; Nichols, R.K.; Morton, E.S.; Stutchbury, B.J.M. Population genetics and relatedness in a critically endangered island raptor, Ridgway’s Hawk Buteo ridgwayi. Conserv. Genet. 2013, 14, 559–571. [Google Scholar] [CrossRef]
- Thiel, T.; Whiteman, N.K.; Tirape, A.; Baquero, M.I.; Cedeno, V.; Walsh, T.; Uzcategui, G.J.; Parker, P.G. Characterization of canarypox-like viruses infecting endemic birds in the Galapagos Islands. J. Wildl. Dis. 2005, 41, 342–353. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Bowden, T.R.; Boyle, D.B. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Yellow-Eyed Penguin (Megadyptes antipodes). Viruses 2021, 13, 194. [Google Scholar] [CrossRef]
- van Riper, C., III; van Riper, S.G.; Hansen, W.R. Epizootiology and Effect of Avian Pox on Hawaiian Forest Birds. Auk 2002, 119, 929–942. [Google Scholar] [CrossRef]
- Young, L.C.; VanderWerf, E.A. Prevalence of avian pox virus and effect on the fledging success of Laysan Albatross. J. Field Ornithol. 2008, 79, 93–98. [Google Scholar] [CrossRef]
- Bolte, A.L.; Meurer, J.; Kaleta, E.F. Avian host spectrum of avipoxviruses. Avian Pathol. 1999, 28, 415–432. [Google Scholar] [CrossRef]
- van Riper, C.; Forrester, D.J. Avian Pox. In Infectious Diseases of Wild Birds; Thomas, N.J., Hunter, D.B., Atkinson, C.T., Eds.; Wiley Blackwell Publishing: Oxford, UK, 2007; pp. 131–176. [Google Scholar]
- Carulei, O.; Douglass, N.; Williamson, A.-L. Comparative analysis of avian poxvirus genomes, including a novel poxvirus from lesser flamingos (Phoenicopterus minor), highlights the lack of conservation of the central region. BMC Genom. 2017, 18, 947. [Google Scholar] [CrossRef]
- Sarker, S.; Das, S.; Lavers, J.L.; Hutton, I.; Helbig, K.; Imbery, J.; Upton, C.; Raidal, S.R. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.). BMC Genom. 2017, 18, 298. [Google Scholar] [CrossRef]
- Tripathy, D.N.; Schnitzlein, W.M.; Morris, P.J.; Janssen, D.L.; Zuba, J.K.; Massey, G.; Atkinson, C.T. Characterization of poxviruses from forest birds in Hawaii. J. Wildl. Dis. 2000, 36, 225–230. [Google Scholar] [CrossRef]
- Niemeyer, C.; Favero, C.M.; Kolesnikovas, C.K.M.; Bhering, R.C.C.; Brandão, P.; Catão-Dias, J.L. Two different avipoxviruses associated with pox disease in Magellanic penguins (Spheniscus magellanicus) along the Brazilian coast. Avian Pathol. 2013, 42, 546–551. [Google Scholar] [CrossRef]
- Joshi, L.R.; Bauermann, F.V.; Hain, K.S.; Kutish, G.F.; Armién, A.G.; Lehman, C.P.; Neiger, R.; Afonso, C.L.; Tripathy, D.N.; Diel, D.G. Detection of Fowlpox virus carrying distinct genome segments of Reticuloendotheliosis virus. Virus Res. 2019, 260, 53–59. [Google Scholar] [CrossRef]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. The genome of fowlpox virus. J. Virol. 2000, 74, 3815–3831. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Raidal, S.R. Molecular characterisation of a novel pathogenic avipoxvirus from an Australian passerine bird, mudlark (Grallina cyanoleuca). Virology 2021, 554, 66–74. [Google Scholar] [CrossRef]
- Sarker, S.; Batinovic, S.; Talukder, S.; Das, S.; Park, F.; Petrovski, S.; Forwood, J.K.; Helbig, K.J.; Raidal, S.R. Molecular characterisation of a novel pathogenic avipoxvirus from the Australian magpie (Gymnorhina tibicen). Virology 2020, 540, 1–16. [Google Scholar] [CrossRef]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Kutish, G.F.; Rock, D.L. The Genome of Canarypox Virus. J. Virol. 2004, 78, 353–366. [Google Scholar] [CrossRef]
- Offerman, K.; Carulei, O.; van der Walt, A.P.; Douglass, N.; Williamson, A.-L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genom. 2014, 15, 1–17. [Google Scholar] [CrossRef]
- Banyai, K.; Palya, V.; Denes, B.; Glavits, R.; Ivanics, E.; Horvath, B.; Farkas, S.L.; Marton, S.; Balint, A.; Gyuranecz, M.; et al. Unique genomic organization of a novel Avipoxvirus detected in turkey (Meleagris gallopavo). Infect Genet. Evol. 2015, 35, 221–229. [Google Scholar] [CrossRef]
- Gyuranecz, M.; Foster, J.T.; Dán, Á.; Ip, H.S.; Egstad, K.F.; Parker, P.G.; Higashiguchi, J.M.; Skinner, M.A.; Höfle, U.; Kreizinger, Z.; et al. Worldwide Phylogenetic Relationship of Avian Poxviruses. J. Virol. 2013, 87, 4938–4951. [Google Scholar] [CrossRef]
- Sarker, S.; Isberg, S.R.; Milic, N.L.; Lock, P.; Helbig, K.J. Molecular characterization of the first saltwater crocodilepox virus genome sequences from the world’s largest living member of the Crocodylia. Sci. Rep. 2018, 8, 5623. [Google Scholar] [CrossRef]
- Graeme, A.T. Action Plan for Seabird Conservation in New Zealand. Part A: Threatened Seabirds. Threatened Species Occasional Publication, 1170-3709; 16, Biodiversity Recovery Unit, Department of Conservation, Wellington, New Zealand. 2000. Available online: https://www.doc.govt.nz/documents/science-and-technical/tsop16.pdf (accessed on 30 January 2021).
- Woods, R. Results of a Preliminary Disease Survey in Shy Albatross (Thalassarche Cauta Gould 1841) Chicks at Albatross Island, Bass Strait, Tasmania; Woods, R., Ed.; Australian Association of Veterinary Conservation Biologists: Canberra, Australia, 2004; pp. 98–104. [Google Scholar]
- Tompkins, E.M.; Anderson, D.J.; Pabilonia, K.L.; Huyvaert, K.P. Avian Pox Discovered in the Critically Endangered Waved Albatross (Phoebastria irrorata) from the Galápagos Islands, Ecuador. J. Wildl. Dis. 2017, 53, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Kane, O.J.; Uhart, M.M.; Rago, V.; Pereda, A.J.; Smith, J.R.; Van Buren, A.; Clark, J.A.; Boersma, P.D. Avian pox in Magellanic Penguins (Spheniscus magellanicus). J. Wildl. Dis. 2012, 48, 790–794. [Google Scholar] [CrossRef] [PubMed]
- Warner, R.E. The Role of Introduced Diseases in the Extinction of the Endemic Hawaiian Avifauna. Condor 1968, 70, 101–120. [Google Scholar] [CrossRef]
- Annuar, B.O.; Mackenzie, J.S.; Lalor, P.A. Isolation and characterization of avipoxviruses from wild birds in Western Australia. Arch. Virol. 1983, 76, 217–229. [Google Scholar] [CrossRef]
- Aleksandr, K.; Olga, B.; David, W.B.; Pavel, P.; Yana, P.; Svetlana, K.; Alexander, N.; Vladimir, R.; Dmitriy, L.; Alexander, S. Non-vector-borne transmission of lumpy skin disease virus. Sci. Rep. 2020, 10, 7436. [Google Scholar] [CrossRef] [PubMed]
- Kitching, R.P.; Taylor, W.P. Transmission of capripoxvirus. Res. Vet. Sci. 1985, 39, 196–199. [Google Scholar] [CrossRef]
- Nakano, E.; Panicali, D.; Paoletti, E. Molecular genetics of vaccinia virus: Demonstration of marker rescue. Proc. Natl. Acad. Sci. USA 1982, 79, 1593–1596. [Google Scholar] [CrossRef]
- Prideaux, C.T.; Boyle, D.B. Fowlpox virus polypeptides: Sequential appearance and virion associated polypeptides. Arch. Virol. 1987, 96, 185–199. [Google Scholar] [CrossRef]
- Sarker, S.; Roberts, H.K.; Tidd, N.; Ault, S.; Ladmore, G.; Peters, A.; Forwood, J.K.; Helbig, K.; Raidal, S.R. Molecular and microscopic characterization of a novel Eastern grey kangaroopox virus genome directly from a clinical sample. Sci. Rep. 2017, 7, 16472. [Google Scholar] [CrossRef]
- Boyle, D.B.; Bulach, D.M.; Amos-Ritchie, R.; Adams, M.M.; Walker, P.J.; Weir, R. Genomic sequences of Australian blue-tongue virus prototype serotypes reveal global relationships and possible routes of entry into Australia. J. Virol. 2012, 86, 6724–6731. [Google Scholar] [CrossRef][Green Version]
- Sarker, S.; Isberg, S.R.; Athukorala, A.; Mathew, R.; Capati, N.; Haque, M.H.; Helbig, K.J. Characterization of a Complete Genome Sequence of Molluscum Contagiosum Virus from an Adult Woman in Australia. Microbiol. Resour. Announc. 2021, 10, e00939-20. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Yang, Z.; Bruno, D.P.; Martens, C.A.; Porcella, S.F.; Moss, B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 11513. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Hofmann, K.; Stoffel, W. TMBASE—A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef]
- Yang, Z.; Reynolds, S.E.; Martens, C.A.; Bruno, D.P.; Porcella, S.F.; Moss, B. Expression profiling of the intermediate and late stages of poxvirus replication. J. Virol. 2011, 85, 9899–9908. [Google Scholar] [CrossRef]
- Yang, Z.; Maruri-Avidal, L.; Sisler, J.; Stuart, C.A.; Moss, B. Cascade regulation of vaccinia virus gene expression is modulated by multistage promoters. Virology 2013, 447, 213–220. [Google Scholar] [CrossRef]
- Goebel, S.J.; Johnson, G.P.; Perkus, M.E.; Davis, S.W.; Winslow, J.P.; Paoletti, E. The complete DNA sequence of vaccinia virus. Virology 1990, 179, 247–266. [Google Scholar] [CrossRef]
- Maizel, J.V., Jr.; Lenk, R.P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 7665–7669. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Hannon, C.; Athukorala, A.; Bielefeldt-Ohmann, H. Emergence of a Novel Pathogenic Poxvirus Infection in the Endangered Green Sea Turtle (Chelonia mydas) Highlights a Key Threatening Process. Viruses 2021, 13, 219. [Google Scholar] [CrossRef]
- Croville, G.; Le Loc’h, G.; Zanchetta, C.; Manno, M.; Camus-Bouclainville, C.; Klopp, C.; Delverdier, M.; Lucas, M.-N.; Donnadieu, C.; Delpont, M.; et al. Rapid whole-genome based typing and surveillance of avipoxviruses using nanopore sequencing. J. Virol. Methods 2018, 261, 34–39. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Bowden, T.R.; Boyle, D.B. Characterisation of an Australian fowlpox virus carrying a near-full-length provirus of reticuloendotheliosis virus. Arch. Virol. 2021, 166, 1485–1488. [Google Scholar] [CrossRef]
- Afonso, C.L.; Tulman, E.R.; Delhon, G.; Lu, Z.; Viljoen, G.J.; Wallace, D.B.; Kutish, G.F.; Rock, D.L. Genome of Crocodilepox Virus. J. Virol. 2006, 80, 4978–4991. [Google Scholar] [CrossRef]
- Sarker, S.; Isberg, R.S.; Moran, L.J.; Araujo, D.R.; Elliott, N.; Melville, L.; Beddoe, T.; Helbig, J.K. Crocodilepox Virus Evolutionary Genomics Supports Observed Poxvirus Infection Dynamics on Saltwater Crocodile (Crocodylus porosus). Viruses 2019, 11, 1116. [Google Scholar] [CrossRef]


| Avipoxviruses (Abbreviation) | GenBank Accession Numbers | Genome Identity (%) | Genome Length (kbp) | A + T Content (%) | Number of ORFs | References |
|---|---|---|---|---|---|---|
| Albatrosspox virus (ALPV) | MW365933 | 352 | 69.9 | 336 | This study | |
| Penguinpox virus 2 (PEPV2) | MW296038 | 98.92 | 350 | 69.9 | 327 | [19] |
| Shearwaterpox virus 2 (SWPV2) | KX857215 | 95.75 | 351 | 69.8 | 312 | [25] |
| Canarypox virus (CNPV) | AY318871 | 92.71 | 360 | 69.6 | 328 | [32] |
| Mudlarkpox virus (MLPV) | MT978051 | 88.47 | 343 | 70.2 | 352 | [30] |
| Magpiepox virus (MPPV) | MK903864 | 78.75 | 293 | 70.4 | 301 | [31] |
| Shearwaterpox virus 1 (SWPV1) | KX857216 | 61.44 | 327 | 72.4 | 310 | [25] |
| Penguinpox virus (PEPV) | KJ859677 | 49.83 | 307 | 70.5 | 285 | [33] |
| Fowlpox virus (FWPV) | AF198100 | 48.89 | 289 | 69.1 | 260 | [29] |
| Pigeonpox virus (FeP2) | KJ801920 | 47.54 | 282 | 70.5 | 271 | [33] |
| Flamingopox virus (FGPV) | MF678796 | 46.51 | 293 | 70.5 | 285 | [24] |
| Turkeypox virus (TKPV) | KP728110 | 33.39 | 189 | 70.2 | 171 | [34] |
| ALPV Synteny | ALPV Genome Coordinates | SWPV2 Synteny | ALPV AA Size | SWPV2 AA Size | SWPV2 BLAST Hits | ALPV AA Identity (%) Compared to Avipoxviruses | ALPV AA Identity (%) Compared to VACV-Cop | VACV BLAST Hits | Notes |
|---|---|---|---|---|---|---|---|---|---|
| ALPV-001 | 827-1342 | SWPV2-001 | 171 | 171 | SWPV2-001 hypothetical protein | 100 | identical to ALPV-336 | ||
| ALPV-002 | 2271-1645 | SWPV2-002 | 208 | 208 | SWPV2-002 C-type lectin-like protein | 100 | identical to ALPV-335 | ||
| ALPV-003 | 2553-2326 | 75 | 56.1 | SWPV1-002 C-type lectin-like protein, identical to ALPV-334 | |||||
| ALPV-004 | 2679-3347 | SWPV2-003 | 222 | 222 | SWPV2-003 conserved hypothetical protein | 100 | identical to ALPV-333 | ||
| ALPV-005 | 3464-3925 | SWPV2-004 | 153 | 134 | SWPV2-004 conserved hypothetical protein | 86.9 | identical to ALPV-332 | ||
| ALPV-006 | 4899-4042 | SWPV2-005 | 285 | 169 | SWPV2-005 C-type lectin-like protein | 46.9 | |||
| ALPV-007 | 5455-4946 | SWPV2-005 | 169 | 169 | SWPV2-005 C-type lectin-like protein | 100 | |||
| ALPV-008 | 7809-5743 | SWPV2-006 | 688 | 688 | SWPV2-006 ankyrin repeat protein | 100 | 23.9 | B4R | |
| ALPV-009 | 8828-8184 | 214 | 43.1 | SWPV1-006 ankyrin repeat protein | |||||
| ALPV-010 | 11218-9458 | SWPV2-007 | 586 | 586 | SWPV2-007 ankyrin repeat protein | 100 | 28.8 | M1L | |
| ALPV-011 | 11483-12052 | SWPV2-008 | 189 | 189 | SWPV2-008 conserved hypothetical protein | 100 | |||
| ALPV-012 | 12765-12259 | SWPV2-009 | 168 | 168 | SWPV2-009 conserved hypothetical protein | 100 | |||
| ALPV-013 | 14555-13083 | SWPV2-010 | 490 | 490 | SWPV2-010 Ig-like domain protein | 100 | |||
| ALPV-014 | 14719-16368 | SWPV2-011 | 549 | 528 | SWPV2-011 ankyrin repeat protein | 96.2 | 28.7 | M1L | |
| ALPV-015 | 16429-16935 | SWPV2-012 | 168 | 168 | SWPV2-012 C-type lectin-like protein | 100 | 24.0 | A40R | |
| ALPV-016 | 17039-18478 | SWPV2-013 | 479 | 479 | SWPV2-013 ankyrin repeat protein | 100 | 26.7 | B4R | |
| ALPV-017 | 19149-18577 | SWPV2-014 | 190 | 190 | SWPV2-014 IL-10-like protein | 100 | |||
| ALPV-018 | 20576-19266 | SWPV2-015 | 436 | 436 | SWPV2-015 ankyrin repeat protein | 100 | 27.5 | M1L | |
| ALPV-019 | 20767-22026 | SWPV2-016 | 419 | 419 | SWPV2-016 ankyrin repeat protein | 100 | 38.2 | M1L | |
| ALPV-020 | 23769-22162 | SWPV2-017 | 535 | 535 | SWPV2-017 ankyrin repeat protein | 100 | 21.5 | B4R | |
| ALPV-021 | 24886-23810 | SWPV2-018 | 358 | 358 | SWPV2-018 putative serpin | 100 | 27.9 | C12L | |
| ALPV-022 | 26242-24968 | SWPV2-019 | 424 | 424 | SWPV2-019 vaccinia C4L/C10L-like protein | 100- | |||
| ALPV-023 | 26520-27056 | SWPV2-020 | 178 | 178 | SWPV2-020 hypothetical protein | 100 | |||
| ALPV-024 | 28172-27270 | SWPV2-021 | 300 | 300 | SWPV2-021 alpha-SNAP-like protein | 100 | |||
| ALPV-025 | 29443-28295 | SWPV2-022 | 382 | 382 | SWPV2-022 ankyrin repeat protein | 100 | 22.0 | C9L | |
| ALPV-026 | 31392-29512 | SWPV2-023 | 626 | 626 | SWPV2-023 ankyrin repeat protein | 100 | |||
| ALPV-027 | 32608-31511 | SWPV2-024 | 365 | 365 | SWPV2-024 ankyrin repeat protein | 100 | |||
| ALPV-028 | 33146-32718 | SWPV2-025 | 142 | 142 | SWPV2-025 C-type lectin-like protein | 100 | |||
| ALPV-029 | 34224-33202 | SWPV2-026 | 340 | 340 | SWPV2-026 ankyrin repeat protein | 100 | 26.8 | B4R | |
| ALPV-030 | 34487-34287 | 66 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-031 | 34467-34826 | SWPV2-027 | 119 | 119 | SWPV2-027 hypothetical protein | 100 | |||
| ALPV-032 | 35782-35054 | SWPV2-028 | 242 | 242 | SWPV2-028 Ig-like domain putative IFN-gamma binding protein | 100 | |||
| ALPV-033 | 36598-35858 | SWPV2-029 | 246 | 246 | SWPV2-029 Ig-like domain protein | 100 | |||
| ALPV-034 | 38673-36694 | SWPV2-030 | 659 | 659 | SWPV2-030 ankyrin repeat protein | 100 | 23.5 | K1L | |
| ALPV-035 | 39392-38991 | SWPV2-031 | 133 | 133 | SWPV2-031 C-type lectin-like protein | 100 | |||
| ALPV-036 | 39776-39489 | SWPV2-032 | 95 | 95 | SWPV2-032 conserved hypothetical protein | 100 | |||
| ALPV-037 | 39840-40379 | SWPV2-033 | 179 | 179 | SWPV2-033 conserved hypothetical protein | 100 | |||
| ALPV-038 | 41625-40384 | SWPV2-034 | 413 | 413 | SWPV2-034 vaccinia C4L/C10L-like protein | 99.8 | 24.3 | C10L | |
| ALPV-039 | 41743-42726 | SWPV2-035 | 327 | 327 | SWPV2-035 G protein-coupled receptor-like protein | 100 | |||
| ALPV-040 | 44520-42745 | SWPV2-036 | 591 | 591 | SWPV2-036 ankyrin repeat protein | 99.7 | 23.2 | B4R | |
| ALPV-041 | 45885-44593 | SWPV2-037 | 430 | 430 | SWPV2-037 ankyrin repeat protein | 100 | 27.7 | B18R | |
| ALPV-042 | 47751-45934 | SWPV2-038 | 605 | 605 | SWPV2-038 ankyrin repeat protein | 100 | 21.6 | B4R | |
| ALPV-043 | 48462-47857 | SWPV2-039 | 201 | 201 | SWPV2-039 conserved hypothetical protein | 100 | |||
| ALPV-044 | 49946-48504 | SWPV2-040 | 480 | 480 | SWPV2-040 ankyrin repeat protein | 100 | 25.2 | B18R | |
| ALPV-045 | 50212-51210 | SWPV2-041 | 332 | 332 | SWPV2-041 G protein-coupled receptor-like protein | 100- | |||
| ALPV-046 | 52589-51237 | SWPV2-042 | 450 | 450 | SWPV2-042 ankyrin repeat protein | 100 | 29.7 | VACV-Cop-B4R | |
| ALPV-047 | 53030-52656 | SWPV2-043 | 124 | 124 | SWPV2-043 conserved hypothetical protein | 100 | |||
| ALPV-048 | 55603-53198 | SWPV2-044 | 801 | 801 | SWPV2-044 alkaline phosphodiesterase-like protein | 100 | |||
| ALPV-049 | 56143-55691 | SWPV2-045 | 150 | 150 | SWPV2-045 hypothetical protein | 100 | |||
| ALPV-050 | 57255-56197 | SWPV2-046 | 352 | 352 | SWPV2-046 ankyrin repeat protein | 100 | |||
| ALPV-051 | 58528-57302 | SWPV2-047 | 408 | 408 | SWPV2-047 DNase II-like protein | 100 | |||
| ALPV-052 | 59069-58554 | SWPV2-048 | 171 | 171 | SWPV2-048 C-type lectin-like protein | 100 | |||
| ALPV-053 | 59689-59249 | SWPV2-049 | 146 | 146 | SWPV2-049 conserved hypothetical protein | 100 | |||
| ALPV-054 | 60104-59682 | SWPV2-050 | 140 | 140 | SWPV2-050 conserved hypothetical protein | 100 | |||
| ALPV-055 | 60647-60156 | SWPV2-051 | 163 | 163 | SWPV2-051 conserved hypothetical protein | 100 | |||
| ALPV-056 | 61081-60644 | SWPV2-052 | 145 | 145 | SWPV2-052 CNLV056 dUTPase | 100 | 52.8 | F2L | |
| ALPV-057 | 62028-61108 | SWPV2-053 | 306 | 306 | SWPV2-053 putative serpin | 100 | |||
| ALPV-058 | 62601-62059 | SWPV2-054 | 180 | 180 | SWPV2-054 bcl-2 like protein | 100 | |||
| ALPV-059 | 63674-62658 | SWPV2-055 | 338 | 338 | SWPV2-055 putative serpin | 100 | |||
| ALPV-060 | 64555-63743 | SWPV2-056 | 270 | 206 | SWPV2-056 conserved hypothetical protein | 99.5 | |||
| ALPV-061 | 66342-64645 | SWPV2-057 | 565 | 565 | SWPV2-057 DNA ligase | 100 | 46.9 | A50R | |
| ALPV-062 | 67433-66381 | SWPV2-058 | 350 | 350 | SWPV2-058 putative serpin | 100 | 25.0 | K2L | |
| ALPV-063 | 68580-67504 | SWPV2-059 | 358 | 358 | SWPV2-059 hydroxysteroid dehydrogenase-like protein | 100 | 38.2 | A44L | |
| ALPV-064 | 69493-68642 | SWPV2-060 | 283 | 283 | SWPV2-060 TGF-beta-like protein | 100 | |||
| ALPV-065 | 71324-69573 | SWPV2-061 | 583 | 583 | SWPV2-061 semaphorin-like protein | 100 | 31.7 | A39R | |
| ALPV-066 | 71606-71424 | SWPV2-062 | 60 | 399 | SWPV2-062 hypothetical protein | 100 | |||
| ALPV-067 | 71608-71784 | 59 | hypothetical protein, unique to ALPV | ||||||
| ALPV-068 | 72000-71728 | SWPV2-062 | 90 | 399 | SWPV2-062 hypothetical protein | 90.8 | |||
| ALPV-069 | 72255-72082 | SWPV2-063 | 57 | 57 | SWPV2-063 hypothetical protein | 100 | |||
| ALPV-070 | 72415-73188 | SWPV2-064 | 257 | 257 | SWPV2-064 GNS1/SUR4-like protein | 100 | |||
| ALPV-071 | 73281-73748 | SWPV2-065 | 155 | 155 | SWPV2-065 late transcription factor VLTF-2 | 100 | 46.6 | A1L | |
| ALPV-072 | 73765-75420 | SWPV2-066 | 551 | 551 | SWPV2-066 putative rifampicin resistance protein, IMV assembly | 100 | 56.2 | D13L | |
| ALPV-073 | 75452-76321 | SWPV2-067 | 289 | 289 | SWPV2-067 mRNA capping enzyme small subunit | 100 | 57.5 | D12L | |
| ALPV-074 | 76342-77283 | SWPV2-068 | 313 | 132 | SWPV2-068 CC chemokine-like protein | 100 | |||
| ALPV-075 | 77690-77361 | SWPV2-069 | 109 | 109 | SWPV2-069 hypothetical protein | 100 | |||
| ALPV-076 | 77761-79668 | SWPV2-070 | 635 | 635 | SWPV2-070 NPH-I, transcription termination factor | 100 | 60.8 | D11L | |
| ALPV-077 | 80351-79665 | SWPV2-071 | 228 | 228 | SWPV2-071 mutT motif putative gene expression regulator | 100 | 38.0 | D10R | |
| ALPV-078 | 81048-80335 | SWPV2-072 | 237 | 232 | SWPV2-072 mutT motif | 97.9 | 46.3 | D9R | |
| ALPV-079 | 81601-81287 | 104 | 100 | CNPV-077 hypothetical protein | |||||
| ALPV-080 | 81796-82065 | 89 | hypothetical protein, unique to ALPV | ||||||
| ALPV-081 | 82693-82448 | 81 | hypothetical protein, unique to ALPV | ||||||
| ALPV-082 | 83172-82690 | SWPV2-073 | 160 | 160 | SWPV2-073 RNA polymerase subunit RPO18 | 100 | 57.5 | D7R | |
| ALPV-083 | 84332-83508 | SWPV2-074 | 274 | 274 | SWPV2-074 Ig-like domain protein | 100 | |||
| ALPV-084 | 86356-84455 | SWPV2-075 | 633 | 633 | SWPV2-075 early transcription factor small subunit VETFS | 100 | 72.2 | D6R | |
| ALPV-085 | 87561-86560 | SWPV2-076 | 333 | 334 | SWPV2-076 Ig-like domain protein | 98.8 | |||
| ALPV-086 | 90271-87887 | SWPV2-077 | 794 | 794 | SWPV2-077 NTPase, DNA replication | 100 | 54.3 | D5R | |
| ALPV-087 | 91091-90426 | SWPV2-078 | 221 | 221 | SWPV2-078 CC chemokine-like protein | 100 | |||
| ALPV-088 | 91830-91174 | SWPV2-079 | 218 | 218 | SWPV2-079 uracil DNA glycosylase | 99.5 | 54.2 | D4R | |
| ALPV-089 | 92626-91871 | SWPV2-080 | 251 | 303 | SWPV2-080 putative RNA phosphatase | 94.7 | 34.1 | H1L | |
| ALPV-090 | 93861-92674 | 395 | 71.9 | SWPV1-075 conserved hypothetical protein | |||||
| ALPV-091 | 93946-94311 | SWPV2-081 | 121 | 112 | SWPV2-081 TNFR-like protein | 80.8 | 33.9 | B28R | |
| ALPV-092 | 96471-96866 | SWPV2-082 | 131 | 131 | SWPV2-082 putative glutathione peroxidase | 100 | |||
| ALPV-093 | 96891-97193 | SWPV2-083 | 100 | 100 | SWPV2-083 conserved hypothetical protein | 100 | |||
| ALPV-094 | 97677-97198 | SWPV2-084 | 159 | 159 | SWPV2-084 conserved hypothetical protein | 100 | |||
| ALPV-095 | 98047-97664 | SWPV2-085 | 127 | 127 | SWPV2-085 conserved hypothetical protein | 100 | |||
| ALPV-096 | 98384-98133 | SWPV2-086 | 83 | 83 | SWPV2-086 HT motif protein | 100 | |||
| ALPV-097 | 99122-98736 | SWPV2-087 | 128 | 146 | SWPV2-087 conserved hypothetical protein | 87.7 | |||
| ALPV-098 | 100027-99224 | SWPV2-088 | 267 | 267 | SWPV2-088 virion protein | 100 | |||
| ALPV-099 | 100102-100929 | SWPV2-089 | 275 | 275 | SWPV2-089 T10-like protein | 100 | |||
| ALPV-100 | 101074-100937 | SWPV2-090 | 45 | 45 | SWPV2-090 conserved hypothetical protein | 100 | |||
| ALPV-101 | 101313-101056 | SWPV2-091 | 85 | 85 | SWPV2-091 ubiquitin | 100 | |||
| ALPV-102 | 102422-101445 | SWPV2-092 | 325 | 339 | SWPV2-092 conserved hypothetical protein | 95.9 | |||
| ALPV-103 | 102692-102450 | SWPV2-093 | 80 | 80 | SWPV2-093 hypothetical protein | 100 | |||
| ALPV-104 | 103285-102698 | SWPV2-094 | 195 | 195 | SWPV2-094 beta-NGF-like protein | 100 | |||
| ALPV-105 | 103815-103309 | SWPV2-095 | 168 | 168 | SWPV2-095 putative interleukin binding protein | 100 | |||
| ALPV-106 | 104127-103870 | SWPV2-096 | 85 | 85 | SWPV2-096 hypothetical protein | 100 | |||
| ALPV-107 | 104455-104138 | SWPV2-097 | 105 | 105 | SWPV2-097 conserved hypothetical protein | 100 | |||
| ALPV-108 | 105044-104472 | SWPV2-098 | 190 | 190 | SWPV2-098 N1R/p28-like protein | 100 | |||
| ALPV-109 | 105246-105623 | SWPV2-099 | 125 | 125 | SWPV2-099 putative glutaredoxin 2, virion morphogenesis | 100 | 32.2 | G4L | |
| ALPV-110 | 106270-105566 | SWPV2-100 | 234 | 234 | SWPV2-100 putative elongation factor | 100 | |||
| ALPV-111 | 106264-106572 | SWPV2-101 | 102 | 102 | SWPV2-101 conserved hypothetical protein | 100 | |||
| ALPV-112 | 106708-106941 | SWPV2-102 | 77 | 77 | SWPV2-102 hypothetical protein | 100 | |||
| ALPV-113 | 107186-109084 | SWPV2-103 | 632 | 632 | SWPV2-103 putative metalloprotease, virion morphogenesis | 100 | 43.9 | G1L | |
| ALPV-114 | 111113-109068 | SWPV2-104 | 681 | 681 | SWPV2-104 NPH-II, RNA helicase | 100 | 43.8 | I8R | |
| ALPV-115 | 111148-112416 | SWPV2-105 | 422 | 422 | SWPV2-105 virion core proteinase | 100 | 54.4 | I7L | |
| ALPV-116 | 112421-113596 | SWPV2-106 | 391 | 391 | SWPV2-106 DNA-binding protein | 100 | 34.8 | I6L | |
| ALPV-117 | 113597-113842 | SWPV2-107 | 81 | 81 | SWPV2-107 putative IMV membrane protein | 100 | |||
| ALPV-118 | 113864-114403 | SWPV2-108 | 179 | 179 | SWPV2-108 thymidine kinase | 100 | 52.0 | J2R | |
| ALPV-119 | 114524-114772 | SWPV2-109 | 82 | 82 | SWPV2-109 HT motif protein | 100 | |||
| ALPV-120 | 114842-115711 | SWPV2-110 | 289 | 289 | SWPV2-110 DNA-binding phosphoprotein | 99.7 | 33.5 | I3L | |
| ALPV-121 | 115712-115921 | SWPV2-111 | 69 | 69 | SWPV2-111 conserved hypothetical protein | 100 | |||
| ALPV-122 | 115928-116860 | SWPV2-112 | 310 | 310 | SWPV2-112 DNA-binding virion protein | 100 | 58.0 | I1L | |
| ALPV-123 | 117040-118998 | SWPV2-113 | 652 | 652 | SWPV2-113 conserved hypothetical protein | 100 | 20.9 | O1L | |
| ALPV-124 | 118928-119323 | SWPV2-114 | 131 | 131 | SWPV2-114 virion core protein | 100 | 34.98 | E11L | |
| ALPV-125 | 119601-119320 | SWPV2-115 | 93 | 93 | SWPV2-115 putative IMV redox protein, virus assembly | 100 | 51.6 | E10R | |
| ALPV-126 | 119628-122594 | SWPV2-116 | 988 | 988 | SWPV2-116 DNA polymerase | 100 | 50.3 | E9L | |
| ALPV-127 | 123413-122586 | 275 | 80.4 | 48.2 | E8R | SWPV1-111 putative membrane protein | |||
| ALPV-128 | 125130-123415 | SWPV2-117 | 571 | 502 | SWPV2-117 conserved hypothetical protein | 87.2 | 49.9 | E6R | |
| ALPV-129 | 130930-125192 | SWPV2-118 | 1912 | 1916 | SWPV2-118 variola B22R-like protein | 99.8 | |||
| ALPV-130 | 136264-130997 | SWPV2-119 | 1755 | 1767 | SWPV2-119 variola B22R-like protein | 99.3 | |||
| ALPV-131 | 142243-136544 | SWPV2-120 | 1899 | 1839 | SWPV2-120 variola B22R-like protein | 95.5 | |||
| ALPV-132 | 142444-142992 | SWPV2-122 | 182 | 182 | SWPV2-122 RNA polymerase subunit RPO30 | 100 | 57.0 | E4L | |
| ALPV-133 | 143024-145189 | SWPV2-123 | 721 | 721 | SWPV2-123 conserved hypothetical protein | 100 | 28.0 | E2L | |
| ALPV-134 | 145182-146600 | SWPV2-124 | 472 | 472 | SWPV2-124 poly(A) polymerase large subunit PAPL | 100 | 50.5 | E1L | |
| ALPV-135 | 146953-146594 | SWPV2-125 | 119 | 119 | SWPV2-125 DNA-binding virion core protein | 100 | 38.8 | F17R | |
| ALPV-136 | 147029-147652 | SWPV2-126 | 207 | 207 | SWPV2-126 conserved hypothetical protein | 100 | |||
| ALPV-137 | 147746-148192 | SWPV2-127 | 148 | 148 | SWPV2-127 conserved hypothetical protein | 100 | 40.4 | F15L | |
| ALPV-138 | 148426-148725 | SWPV2-128 | 99 | 99 | SWPV2-128 conserved hypothetical protein | 100 | |||
| ALPV-139 | 154234-148796 | SWPV2-129 | 1812 | 1801 | SWPV2-129 variola B22R-like protein | 99.4 | |||
| ALPV-140 | 154384-155520 | SWPV2-130 | 378 | 378 | SWPV2-130 putative palmitoylated EEV envelope lipase | 100 | 38.0 | F13L | |
| ALPV-141 | 155598-157475 | SWPV2-131 | 625 | 625 | SWPV2-131 putative EEV maturation protein | 100 | 26.3 | F12L | |
| ALPV-142 | 157518-158906 | SWPV2-132 | 462 | 462 | SWPV2-132 conserved hypothetical protein | 100 | 27.0 | F11L | |
| ALPV-143 | 158997-160331 | SWPV2-133 | 444 | 444 | SWPV2-133 putative serine/threonine protein kinase, virus assembly | 100 | 53.6 | F10L | |
| ALPV-144 | 160306-160947 | SWPV2-134 | 213 | 213 | SWPV2-134 conserved hypothetical protein | 100 | 31.5 | F9L | |
| ALPV-145 | 161030-161230 | SWPV2-135 | 66 | 66 | SWPV2-135 conserved hypothetical protein | 100 | |||
| ALPV-146 | 161556-162110 | SWPV2-136 | 184 | 184 | SWPV2-136 HAL3-like domain protein | 100 | |||
| ALPV-147 | 162371-163336 | SWPV2-137 | 321 | 321 | SWPV2-137 N1R/p28-like protein | 100 | |||
| ALPV-148 | 163448-165463 | SWPV2-138 | 671 | 671 | SWPV2-138 ankyrin repeat protein | 100 | 26.0 | M1L | |
| ALPV-149 | 165489-167159 | SWPV2-139 | 556 | 556 | SWPV2-139 ankyrin repeat protein | 100 | 26.1 | B4R | |
| ALPV-150 | 167380-168702 | SWPV2-140 | 440 | 440 | SWPV2-140 conserved hypothetical protein | 100 | 33.3 | G5R | |
| ALPV-151 | 168710-168898 | SWPV2-141 | 62 | 62 | SWPV2-141 RNA polymerase subunit RPO7 | 98.4 | 55.2 | G5.5R | |
| ALPV-152 | 168891-169457 | SWPV2-142 | 188 | 188 | SWPV2-142 conserved hypothetical protein | 100 | 31.8 | G6R | |
| ALPV-153 | 170468-169422 | SWPV2-143 | 348 | 348 | SWPV2-143 virion core protein | 100 | 34.6 | G7L | |
| ALPV-154 | 171554-170634 | SWPV2-144 | 306 | 306 | SWPV2-144 putative thioredoxin binding protein | 100 | |||
| ALPV-155 | 171682-171915 | 77 | 97.1 | CNPV-150 ankyrin repeat protein | |||||
| ALPV-156 | 173269-172031 | SWPV2-145 | 412 | 412 | SWPV2-145 ankyrin repeat protein | 100 | 42.9 | M1L | |
| ALPV-157 | 173945-173496 | SWPV2-146 | 149 | 149 | SWPV2-146 hypothetical protein | 100 | |||
| ALPV-158 | 175111-174173 | SWPV2-147 | 312 | 312 | SWPV2-147 Rep-like protein | 100 | |||
| ALPV-159 | 181355-175545 | SWPV2-148 | 1936 | 875 | SWPV2-148 variola B22R-like protein | 98.2 | |||
| ALPV-160 | 186885-181408 | SWPV2-149 | 1825 | 1831 | SWPV2-149 variola B22R-like protein | 99.7 | |||
| ALPV-161 | 187202-189685 | SWPV2-150 | 827 | 834 | SWPV2-150 hypothetical protein | 90.5 | |||
| ALPV-162 | 190813-189782 | SWPV2-151 | 343 | 343 | SWPV2-151 TGF-beta-like protein | 100 | |||
| ALPV-163 | 190815-191294 | 159 | 64.7 | CNPV-157 TGF-beta-like protein | |||||
| ALPV-164 | 191763-192263 | SWPV2-150 | 166 | 834 | SWPV2-150 hypothetical protein | 47.8 | |||
| ALPV-165 | 192733-193446 | 237 | 97.2 | CNPV-159 N1R/p28-like protein | |||||
| ALPV-166 | 193490-194494 | 334 | 91.3 | CNPV-169 N1R/p28-like protein | |||||
| ALPV-167 | 194549-195412 | 287 | 78.7 | CNPV-169 N1R/p28-like protein | |||||
| ALPV-168 | 195417-195821 | 134 | 98.5 | CNPV-160 N1R/p28-like protein | |||||
| ALPV-169 | 197008-195920 | SWPV2-152 | 362 | 358 | SWPV2-152 TGF-beta-like protein | 98.9 | |||
| ALPV-170 | 197058-197507 | SWPV2-153 | 149 | 149 | SWPV2-153 TGF-beta-like protein | 100 | |||
| ALPV-171 | 197843-198883 | SWPV2-154 | 346 | 320 | SWPV2-154 N1R/p28-like protein | 95.3 | |||
| ALPV-172 | 199118-200155 | SWPV2-155 | 345 | 345 | SWPV2-155 Ig-like domain protein | 99.7 | |||
| ALPV-173 | 200427-200933 | SWPV2-156 | 168 | 168 | SWPV2-156 Ig-like domain protein | 97 | |||
| ALPV-174 | 201028-202059 | SWPV2-157 | 343 | 350 | SWPV2-157 N1R/p28-like protein | 93.6 | |||
| ALPV-175 | 202011-202415 | 134 | 92.5 | CNPV-226 N1R/p28-like protein | |||||
| ALPV-176 | 202488-203126 | SWPV2-158 | 212 | 212 | SWPV2-158 thymidylate kinase | 100 | 45.2 | A48R | |
| ALPV-177 | 203179-203961 | SWPV2-159 | 260 | 260 | SWPV2-159 late transcription factor VLTF-1 | 100 | 66.2 | G8R | |
| ALPV-178 | 203975-204982 | SWPV2-160 | 335 | 335 | SWPV2-160 putative myristylated protein | 100 | 38.0 | G9R | |
| ALPV-179 | 204983-205714 | SWPV2-161 | 243 | 243 | SWPV2-161 putative myristylated IMV envelope protein | 100 | 54.7 | L1R | |
| ALPV-180 | 205774-206064 | SWPV2-162 | 96 | 96 | SWPV2-162 conserved hypothetical protein | 100 | |||
| ALPV-181 | 206965-206054 | SWPV2-163 | 303 | 303 | SWPV2-163 conserved hypothetical protein | 100 | 42.0 | L3L | |
| ALPV-182 | 206991-207749 | SWPV2-164 | 252 | 252 | SWPV2-164 DNA-binding virion core protein | 100 | 36.3 | L4R | |
| ALPV-183 | 207750-208142 | SWPV2-165 | 130 | 130 | SWPV2-165 conserved hypothetical protein | 100 | 41.6 | L5R | |
| ALPV-184 | 208096-208542 | SWPV2-166 | 148 | 148 | SWPV2-166 putative IMV membrane protein | 100 | 42.5 | J1R | |
| ALPV-185 | 208576-209484 | SWPV2-167 | 302 | 302 | SWPV2-167 poly(A) polymerase small subunit PAPS | 100 | 56.0 | J3R | |
| ALPV-186 | 209481-210041 | SWPV2-168 | 186 | 186 | SWPV2-168 RNA polymerase subunit RPO22 | 100 | 55.0 | J4R | |
| ALPV-187 | 210444-210034 | SWPV2-169 | 136 | 136 | SWPV2-169 conserved hypothetical protein | 100 | 47.9 | J5L | |
| ALPV-188 | 210487-214353 | SWPV2-170 | 1288 | 1288 | SWPV2-170 RNA polymerase subunit RPO147 | 100 | 70.6 | J6R | |
| ALPV-189 | 214856-214356 | SWPV2-171 | 166 | 166 | SWPV2-171 putative protein-tyrosine phosphatase, virus assembly | 100 | 47.6 | H1L | |
| ALPV-190 | 214872-215441 | SWPV2-172 | 189 | 189 | SWPV2-172 conserved hypothetical protein | 100 | 48.9 | H2R | |
| ALPV-191 | 216503-215517 | SWPV2-173 | 328 | 328 | SWPV2-173 ankyrin repeat protein | 100 | |||
| ALPV-192 | 217538-216546 | SWPV2-174 | 330 | 330 | SWPV2-174 putative IMV envelope protein | 100 | 32.2 | H3L | |
| ALPV-193 | 220029-217630 | SWPV2-175 | 799 | 799 | SWPV2-175 RNA polymerase associated protein RAP94 | 100 | 55.2 | H4L | |
| ALPV-194 | 220198-220710 | SWPV2-176 | 170 | 170 | SWPV2-176 late transcription factor VLTF-4 | 100 | |||
| ALPV-195 | 220711-221661 | SWPV2-177 | 316 | 316 | SWPV2-177 DNA topoisomerase | 100 | 58.0 | H6R | |
| ALPV-196 | 221666-222127 | SWPV2-178 | 153 | 153 | SWPV2-178 conserved hypothetical protein | 100 | 33.8 | H7R | |
| ALPV-197 | 222401-222090 | SWPV2-179 | 103 | 103 | SWPV2-179 conserved hypothetical protein | 100 | |||
| ALPV-198 | 222409-224949 | SWPV2-180 | 846 | 846 | SWPV2-180 mRNA capping enzyme large subunit | 100 | 53.9 | D1R | |
| ALPV-199 | 225020-225340 | SWPV2-181 | 106 | 106 | SWPV2-181 HT motif protein | 100 | |||
| ALPV-200 | 225759-225337 | SWPV2-182 | 140 | 140 | SWPV2-182 virion protein | 100 | |||
| ALPV-201 | 225813-226247 | SWPV2-183 | 144 | 144 | SWPV2-183 hypothetical protein | 100 | |||
| ALPV-202 | 226312-226884 | SWPV2-184 | 190 | 190 | SWPV2-184 conserved hypothetical protein | 100 | |||
| ALPV-203 | 226950-227777 | SWPV2-185 | 275 | 275 | SWPV2-185 N1R/p28-like protein | 100 | |||
| ALPV-204 | 228314-227844 | SWPV2-186 | 156 | 156 | SWPV2-186 C-type lectin-like protein | 100 | 30.8 | A34R | |
| ALPV-205 | 228622-229299 | SWPV2-187 | 225 | 225 | SWPV2-187 deoxycytidine kinase-like protein | 100 | |||
| ALPV-206 | 229305-229805 | SWPV2-188 | 166 | 166 | SWPV2-188 Rep-like protein | 99.4 | |||
| ALPV-207 | 229864-230367 | SWPV2-189 | 167 | 167 | SWPV2-189 conserved hypothetical protein | 100 | |||
| ALPV-208 | 230421-231251 | SWPV2-190 | 276 | 276 | SWPV2-190 N1R/p28-like protein | 100 | |||
| ALPV-209 | 231324-232472 | SWPV2-191 | 382 | 382 | SWPV2-191 N1R/p28-like protein | 100 | |||
| ALPV-210 | 232528-232713 | SWPV2-192 | 61 | 61 | SWPV2-192 conserved hypothetical protein | 100 | |||
| ALPV-211 | 232932-233888 | SWPV2-193 | 318 | 318 | SWPV2-193 N1R/p28-like protein | 100 | |||
| ALPV-212 | 233949-235367 | SWPV2-194 | 472 | 472 | SWPV2-194 putative photolyase | 100 | |||
| ALPV-213 | 235424-235579 | 51 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-214 | 235557-236078 | SWPV2-195 | 173 | 173 | SWPV2-195 N1R/p28-like protein | 100 | |||
| ALPV-215 | 236184-236723 | SWPV2-196 | 179 | 200 | SWPV2-196 conserved hypothetical protein | 89 | |||
| ALPV-216 | 236767-237699 | SWPV2-197 | 310 | 310 | SWPV2-197 N1R/p28-like protein | 100 | |||
| ALPV-217 | 237747-238142 | SWPV2-198 | 131 | 131 | SWPV2-198 N1R/p28-like protein | 100 | |||
| ALPV-218 | 238197-238361 | SWPV2-199 | 54 | 54 | SWPV2-199 conserved hypothetical protein | 100 | |||
| ALPV-219 | 238421-238951 | SWPV2-200 | 176 | 176 | SWPV2-200 N1R/p28-like protein | 100 | |||
| ALPV-220 | 239662-239012 | SWPV2-201 | 216 | 216 | SWPV2-201 deoxycytidine kinase-like protein | 100 | |||
| ALPV-221 | 239836-240906 | SWPV2-202 | 356 | 356 | SWPV2-202 vaccinia C4L/C10L-like protein | 100 | 28.1 | C10L | |
| ALPV-222 | 241181-241795 | SWPV2-203 | 204 | 204 | SWPV2-203 CC chemokine-like protein | 100 | |||
| ALPV-223 | 241885-243090 | SWPV2-204 | 401 | 401 | SWPV2-204 conserved hypothetical protein | 100 | |||
| ALPV-224 | 243204-244196 | SWPV2-205 | 330 | 330 | SWPV2-205 N1R/p28-like protein | 100 | |||
| ALPV-225 | 244284-245555 | SWPV2-206 | 423 | 223 | SWPV2-206 N1R/p28-like protein | 99.5 | |||
| ALPV-226 | 245795-245619 | 58 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-227 | 246013-245858 | 51 | hypothetical protein, unique to ALPV, containing a transmembrane helix | ||||||
| ALPV-228 | 246363-247412 | SWPV2-207 | 349 | 349 | SWPV2-207 N1R/p28-like protein | 100 | |||
| ALPV-229 | 247466-248308 | 280 | 80.9 | SWPV1-198 N1R/p28-like protein | |||||
| ALPV-230 | 248847-248644 | SWPV2-208 | 67 | 85 | SWPV2-208 N1R/p28-like protein | 62.7 | |||
| ALPV-231 | 249295-249483 | SWPV2-209 | 62 | 213 | SWPV2-209 N1R/p28-like protein | 100 | |||
| ALPV-232 | 250029-250292 | SWPV2-210 | 87 | 285 | SWPV2-210 N1R/p28-like protein | 100 | |||
| ALPV-233 | 250283-250615 | 110 | 98.2 | CNPV-227 N1R/p28-like protein | |||||
| ALPV-234 | 253677-251134 | SWPV2-211 | 847 | 847 | SWPV2-211 ankyrin repeat protein | 99.9 | 31.6 | B4R | |
| ALPV-235 | 253931-254650 | SWPV2-212 | 239 | 239 | SWPV2-212 hypothetical protein | 100 | |||
| ALPV-236 | 254650-255027 | 125 | 73.3 | CNPV-227 N1R/p28-like protein | |||||
| ALPV-237 | 255062-255346 | SWPV2-214 | 94 | 126 | SWPV2-214 N1R/p28-like protein | 96.4 | |||
| ALPV-238 | 257103-255799 | SWPV2-215 | 434 | 434 | SWPV2-215 ankyrin repeat protein | 100 | 27.3 | B4R | |
| ALPV-239 | 257301-257498 | SWPV2-216 | 65 | 65 | SWPV2-216 hypothetical protein | 100 | |||
| ALPV-240 | 257446-257922 | SWPV2-217 | 158 | 158 | SWPV2-217 MyD116-like domain protein | 100 | |||
| ALPV-241 | 257952-258566 | SWPV2-218 | 204 | 204 | SWPV2-218 CC chemokine-like protein | 98 | |||
| ALPV-242 | 258748-260163 | SWPV2-219 | 471 | 471 | SWPV2-219 ankyrin repeat protein | 100 | 31.4 | M1L | |
| ALPV-243 | 260183-261709 | SWPV2-220 | 508 | 508 | SWPV2-220 ankyrin repeat protein | 100 | 37.0 | M1L | |
| ALPV-244 | 261780-263084 | SWPV2-221 | 434 | 432 | SWPV2-221 conserved hypothetical protein | 99.5 | |||
| ALPV-245 | 263129-264100 | SWPV2-222 | 323 | 323 | SWPV2-222 ribonucleotide reductase small subunit | 100 | 70.9 | F4L | |
| ALPV-246 | 264281-265606 | SWPV2-223 | 441 | 441 | SWPV2-223 ankyrin repeat protein | 99.8 | 33.3 | B4R | |
| ALPV-247 | 266342-265665 | SWPV2-224 | 225 | 225 | SWPV2-224 late transcription factor VLTF-3 | 100 | 76.7 | A2L | |
| ALPV-248 | 266557-266330 | SWPV2-225 | 75 | 75 | SWPV2-225 virion redox protein | 100 | |||
| ALPV-249 | 268550-266571 | SWPV2-226 | 659 | 659 | SWPV2-226 virion core protein P4b | 100 | 54.1 | A3L | |
| ALPV-250 | 269284-268637 | SWPV2-227 | 215 | 215 | SWPV2-227 immunodominant virion protein | 100 | |||
| ALPV-251 | 269323-269832 | SWPV2-228 | 169 | 169 | SWPV2-228 RNA polymerase subunit RPO19 | 100 | 55.2 | A5R | |
| ALPV-252 | 270948-269827 | SWPV2-229 | 373 | 373 | SWPV2-229 conserved hypothetical protein | 100 | 39.3 | A6L | |
| ALPV-253 | 273084-270955 | SWPV2-230 | 709 | 709 | SWPV2-230 early transcription factor large subunit VETFL | 100 | 59.6 | A7L | |
| ALPV-254 | 273148-274050 | SWPV2-231 | 300 | 300 | SWPV2-231 intermediate transcription factor VITF-3 | 100 | 38.7 | A8R | |
| ALPV-255 | 274242-274015 | SWPV2-232 | 75 | 75 | SWPV2-232 putative IMV membrane protein | 100 | |||
| ALPV-256 | 276924-274243 | SWPV2-233 | 893 | 893 | SWPV2-233 virion core protein P4a | 100 | 39.6 | A10L | |
| ALPV-257 | 276942-277781 | SWPV2-234 | 279 | 279 | SWPV2-234 conserved hypothetical protein | 100 | 39.5 | A11R | |
| ALPV-258 | 278284-277778 | SWPV2-235 | 168 | 168 | SWPV2-235 virion protein | 99.4 | 32.9 | A12L | |
| ALPV-259 | 278299-278556 | SWPV2-236 | 85 | 56 | SWPV2-236 conserved hypothetical protein | 87.0 | |||
| ALPV-260 | 278754-278545 | SWPV2-237 | 69 | 69 | SWPV2-237 putative IMV membrane protein | 100 | |||
| ALPV-261 | 279080-278802 | SWPV2-238 | 92 | 92 | SWPV2-238 putative IMV membrane protein | 100 | 27.5 | A14L | |
| ALPV-262 | 279258-279097 | SWPV2-239 | 53 | 53 | SWPV2-239 putative IMV membrane virulence factor | 100 | 35.3 | A 14.5L | |
| ALPV-263 | 279564-279274 | SWPV2-240 | 96 | 96 | SWPV2-240 conserved hypothetical protein | 100 | |||
| ALPV-264 | 280654-279548 | SWPV2-241 | 368 | 368 | SWPV2-241 predicted myristylated protein | 100 | 43.0 | A16L | |
| ALPV-265 | 281248-280670 | SWPV2-242 | 192 | 192 | SWPV2-242 putative phosphorylated IMV membrane protein | 100 | 33.2 | A17L | |
| ALPV-266 | 281266-282654 | SWPV2-243 | 462 | 462 | SWPV2-243 DNA helicase, transcriptional elongation | 100 | 50.2 | A18R | |
| ALPV-267 | 282891-282622 | SWPV2-244 | 89 | 89 | SWPV2-244 conserved hypothetical protein | 100 | 42.9 | A19L | |
| ALPV-268 | 283237-282899 | SWPV2-245 | 112 | 112 | SWPV2-245 conserved hypothetical protein | 100 | 45.3 | A21L | |
| ALPV-269 | 283236-284540 | SWPV2-246 | 434 | 434 | SWPV2-246 DNA polymerase processivity factor | 100 | 25.9 | A20R | |
| ALPV-270 | 284537-284995 | SWPV2-247 | 152 | 152 | SWPV2-247 Holliday junction resolvase protein | 100 | 45.0 | A22R | |
| ALPV-271 | 285012-286163 | SWPV2-248 | 383 | 383 | SWPV2-248 intermediate transcription factor VITF-3 | 100 | 51.3 | A23R | |
| ALPV-272 | 286189-289662 | SWPV2-249 | 1157 | 1157 | SWPV2-249 RNA polymerase subunit RPO132 | 100 | 74.6 | A24R | |
| ALPV-273 | 291456-289651 | SWPV2-250 | 601 | 601 | SWPV2-250 A type inclusion-like protein | 100 | |||
| ALPV-274 | 292918-291491 | SWPV2-251 | 475 | 475 | SWPV2-251 A type inclusion-like/fusion protein | 100 | 56.4 | A27L | |
| ALPV-275 | 293341-292919 | SWPV2-252 | 140 | 140 | SWPV2-252 conserved hypothetical protein | 100 | 41.8 | A28L | |
| ALPV-276 | 294263-293346 | SWPV2-253 | 305 | 305 | SWPV2-253 RNA polymerase subunit RPO35 | 100 | 42.6 | A29L | |
| ALPV-277 | 294465-294238 | SWPV2-254 | 75 | 75 | SWPV2-254 conserved hypothetical protein | 100 | |||
| ALPV-278 | 294590-294931 | SWPV2-255 | 113 | 113 | SWPV2-255 conserved hypothetical protein | 100 | 31.6 | A31R | |
| ALPV-279 | 294940-295302 | SWPV2-256 | 120 | 120 | SWPV2-256 conserved hypothetical protein | 100 | |||
| ALPV-280 | 296145-295291 | SWPV2-257 | 284 | 284 | SWPV2-257 DNA packaging protein | 100 | 47.1 | A32L | |
| ALPV-281 | 296260-296805 | SWPV2-258 | 181 | 181 | SWPV2-258 C-type lectin-like EEV protein | 100 | |||
| ALPV-282 | 297030-297854 | SWPV2-259 | 274 | 274 | SWPV2-259 conserved hypothetical protein | 100 | |||
| ALPV-283 | 297914-298723 | SWPV2-260 | 269 | 269 | SWPV2-260 putative tyrosine protein kinase | 100 | |||
| ALPV-284 | 298766-299782 | SWPV2-261 | 338 | 338 | SWPV2-261 putative serpin | 100 | |||
| ALPV-285 | 300562-299804 | SWPV2-262 | 252 | 252 | SWPV2-262 conserved hypothetical protein | 100 | |||
| ALPV-286 | 300672-301604 | SWPV2-263 | 310 | 310 | SWPV2-263 G protein-coupled receptor-like protein | 100 | |||
| ALPV-287 | 301615-301905 | SWPV2-264 | 96 | 96 | SWPV2-264 conserved hypothetical protein | 100 | |||
| ALPV-288 | 301971-302501 | SWPV2-265 | 176 | 169 | SWPV2-265 beta-NGF-like protein | 96 | |||
| ALPV-289 | 302911-302519 | SWPV2-266 | 130 | 130 | SWPV2-266 HT motif protein | 100 | |||
| ALPV-290 | 303015-303659 | SWPV2-267 | 214 | 214 | SWPV2-267 conserved hypothetical protein | 100 | |||
| ALPV-291 | 304032-303670 | SWPV2-268 | 120 | 120 | SWPV2-268 HT motif protein | 100 | |||
| ALPV-292 | 304198-304533 | SWPV2-269 | 111 | 111 | SWPV2-269 CC chemokine-like protein | 100 | |||
| ALPV-293 | 304612-305193 | SWPV2-270 | 193 | 193 | SWPV2-270 putative interleukin binding protein | 100 | |||
| ALPV-294 | 305303-305683 | SWPV2-271 | 126 | 126 | SWPV2-271 EGF-like protein | 100 | 34.8 | C11R | |
| ALPV-295 | 305685-306602 | SWPV2-272 | 305 | 305 | SWPV2-272 putative serine/threonine protein kinase | 100 | 41.1 | B1R | |
| ALPV-296 | 306645-307127 | SWPV2-273 | 160 | 160 | SWPV2-273 conserved hypothetical protein | 100 | |||
| ALPV-297 | 307210-307677 | SWPV2-274 | 155 | 147 | SWPV2-274 C-type lectin-like protein | 99.3 | 22.6 | A34R | |
| ALPV-298 | 307720-308139 | SWPV2-275 | 139 | 139 | SWPV2-275 putative interleukin binding protein | 100 | |||
| ALPV-299 | 308208-308435 | SWPV2-276 | 75 | 75 | SWPV2-276 conserved hypothetical protein | 100 | |||
| ALPV-300 | 308637-310421 | SWPV2-277 | 594 | 594 | SWPV2-277 ankyrin repeat protein | 99.8 | 22.3 | B4R | |
| ALPV-301 | 310445-310669 | SWPV2-278 | 74 | 74 | SWPV2-278 hypothetical protein | 100 | |||
| ALPV-302 | 310712-311566 | SWPV2-279 | 284 | 284 | SWPV2-279 ankyrin repeat protein | 99.6 | 33.9 | B24R | |
| ALPV-303 | 311621-312913 | SWPV2-280 | 430 | 430 | SWPV2-280 ankyrin repeat protein | 99.8 | C18L | ||
| ALPV-304 | 313106-314296 | SWPV2-281 | 396 | 396 | SWPV2-281 ankyrin repeat protein | 100 | 28.6 | M1L | |
| ALPV-305 | 314299-315675 | SWPV2-282 | 458 | 458 | SWPV2-282 ankyrin repeat protein | 100 | 32.5 | B4R | |
| ALPV-306 | 315784-317997 | SWPV2-283 | 737 | 737 | SWPV2-283 ankyrin repeat protein | 100 | 25.8 | M1L | |
| ALPV-307 | 318053-319768 | SWPV2-284 | 571 | 571 | SWPV2-284 ankyrin repeat protein | 100 | 28.8 | B4R | |
| ALPV-308 | 319772-320674 | SWPV2-285 | 300 | 300 | SWPV2-285 putative serine/threonine protein kinase | 100 | 24.6 | M1L | |
| ALPV-309 | 320747-321481 | SWPV2-286 | 244 | 244 | SWPV2-286 ankyrin repeat protein | 100 | 32.8 | B1R | |
| ALPV-310 | 322082-323665 | SWPV2-287 | 527 | 527 | SWPV2-287 ankyrin repeat protein | 100 | 23.7 | M1L | |
| ALPV-311 | 324261-323680 | SWPV2-288 | 193 | 193 | SWPV2-288 conserved hypothetical protein | 100 | 26.1 | B18R | |
| ALPV-312 | 324329-325831 | SWPV2-289 | 500 | 500 | SWPV2-289 ankyrin repeat protein | 100 | |||
| ALPV-313 | 326047-327447 | SWPV2-290 | 466 | 466 | SWPV2-290 ankyrin repeat protein | 100 | 28.3 | M1L | |
| ALPV-314 | 327518-328306 | SWPV2-291 | 262 | 262 | SWPV2-291 N1R/p28-like protein | 100 | 28.2 | B4R | |
| ALPV-315 | 328368-328586 | SWPV2-292 | 72 | 72 | SWPV2-292 hypothetical protein | 100 | |||
| ALPV-316 | 329054-328590 | SWPV2-293 | 154 | 154 | SWPV2-293 C-type lectin-like protein | 100 | |||
| ALPV-317 | 329231-330304 | SWPV2-294 | 357 | 357 | SWPV2-294 ankyrin repeat protein | 100 | 34.4 | A40R | |
| ALPV-318 | 330452-331042 | SWPV2-295 | 196 | 196 | SWPV2-295 ankyrin repeat protein | 100 | |||
| ALPV-319 | 331147-332760 | SWPV2-296 | 537 | 537 | SWPV2-296 ankyrin repeat protein | 100 | 37.4 | M1L | |
| ALPV-320 | 332794-333168 | SWPV2-297 | 124 | 124 | SWPV2-297 EFc-like protein | 100 | |||
| ALPV-321 | 333178-333678 | SWPV2-298 | 166 | 166 | SWPV2-298 conserved hypothetical protein | 100 | |||
| ALPV-322 | 333750-334406 | SWPV2-299 | 218 | 218 | SWPV2-299 Ig-like domain protein | 100 | |||
| ALPV-323 | 334433-336322 | SWPV2-300 | 629 | 629 | SWPV2-300 ankyrin repeat protein | 99.8 | |||
| ALPV-324 | 336421-337368 | SWPV2-301 | 315 | 315 | SWPV2-301 G protein-coupled receptor-like protein | 100 | 40.8 | B4R | |
| ALPV-325 | 337435-339069 | SWPV2-302 | 544 | 544 | SWPV2-302 ankyrin repeat protein | 99.8 | |||
| ALPV-326 | 339250-339417 | SWPV2-303 | 55 | 55 | SWPV2-303 hypothetical protein | 100 | 33.9 | B4R | |
| ALPV-327 | 339592-341124 | SWPV2-304 | 510 | 514 | SWPV2-304 ankyrin repeat protein | 99.2 | |||
| ALPV-328 | 341457-343424 | SWPV2-305 | 655 | 637 | SWPV2-305 ankyrin repeat protein | 96.6 | 36.8 | M1L | |
| ALPV-329 | 343616-345025 | SWPV2-306 | 469 | 469 | SWPV2-306 Ig-like domain protein | 100 | 25.8 | M1L | |
| ALPV-330 | 345157-345531 | SWPV2-307 | 124 | 124 | SWPV2-307 EFc-like protein | 100 | |||
| ALPV-331 | 345865-347868 | SWPV2-308 | 667 | 689 | SWPV2-308 ankyrin repeat protein | 96.5 | |||
| ALPV-332 | 348446-347985 | SWPV2-309 | 153 | 186 | SWPV2-309 conserved hypothetical protein | 86.9 | identical to ALPV-005 | ||
| ALPV-333 | 349231-348563 | SWPV2-310 | 222 | 222 | SWPV2-310 conserved hypothetical protein | 100 | identical to ALPV-004 | ||
| ALPV-334 | 349357-349584 | 75 | 56.1 | SWPV1-002 C-type lectin-like protein, identical to ALPV-003 | |||||
| ALPV-335 | 349639-350265 | SWPV2-311 | 208 | 208 | SWPV2-311 C-type lectin-like protein | 100 | identical to ALPV-002 | ||
| ALPV-336 | 351083-350568 | SWPV2-312 | 171 | 171 | SWPV2-312 hypothetical protein | 100 | identical to ALPV-001 |
| Virus | Abbreviation | GenBank Accession Number | Reference |
|---|---|---|---|
| Albatrosspox virus | ALPV | MW365933 | This study |
| Canarypox virus | CNPV | AY318871 | [32] |
| Cheloniidpox virus 1 | ChePV-1 | MT799800 | [61] |
| Fowlpox virus | FWPV | AF198100, MF766430-32, MH709124-25, MH719203, MH734528, AJ581527, MW142017 | [28,29,62,63] |
| Flamingopox virus | FGPV | MF678796 | [24] |
| Magpiepox virus | MPPV | MK903864 | [31] |
| Mudlarkpox virus | MLPV | MT978051 | [30] |
| Nile crocodilepox virus | CRV | DQ356948 | [64] |
| Penguinpox virus | PEPV | KJ859677 | [33] |
| Penguinpox virus 2 | PEPV2 | MW296038 | [19] |
| Pigeonpox virus | FeP2 | KJ801920 | [33] |
| Saltwater crocodilepox virus 1 | SwCRV1 | MG450915 | [36,65] |
| Shearwaterpox virus 1 | SWPV1 | KX857216 | [25] |
| Shearwaterpox virus 2 | SWPV2 | KX857215 | [25] |
| Turkeypox virus | TKPV | NC_028238 | [34] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarker, S.; Athukorala, A.; Nyandowe, T.; Bowden, T.R.; Boyle, D.B. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi). Pathogens 2021, 10, 575. https://doi.org/10.3390/pathogens10050575
Sarker S, Athukorala A, Nyandowe T, Bowden TR, Boyle DB. Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi). Pathogens. 2021; 10(5):575. https://doi.org/10.3390/pathogens10050575
Chicago/Turabian StyleSarker, Subir, Ajani Athukorala, Tadiwa Nyandowe, Timothy R. Bowden, and David B. Boyle. 2021. "Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi)" Pathogens 10, no. 5: 575. https://doi.org/10.3390/pathogens10050575
APA StyleSarker, S., Athukorala, A., Nyandowe, T., Bowden, T. R., & Boyle, D. B. (2021). Genomic Characterisation of a Novel Avipoxvirus Isolated from an Endangered Northern Royal Albatross (Diomedea sanfordi). Pathogens, 10(5), 575. https://doi.org/10.3390/pathogens10050575






