Structural Modelling Prediction of Recombinant Plasmodium falciparum K13-F446I and K13-C580Y Gene by AlphaFold Method and Heterologous Expression in Spodoptera frugiperda 9 Cells
Abstract
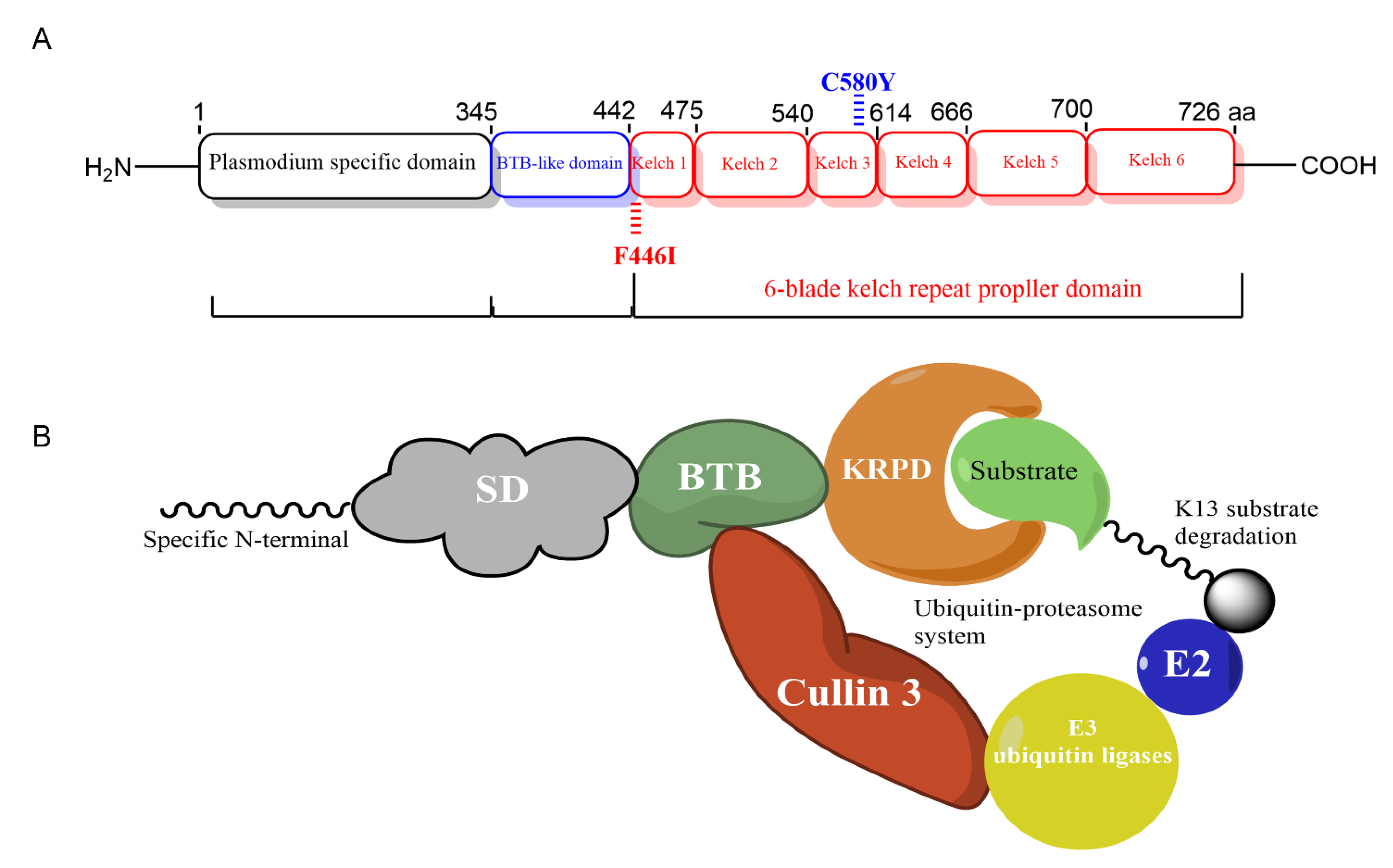
:1. Introduction
2. Materials and Methods
2.1. Strains, Insect Cells and Materials
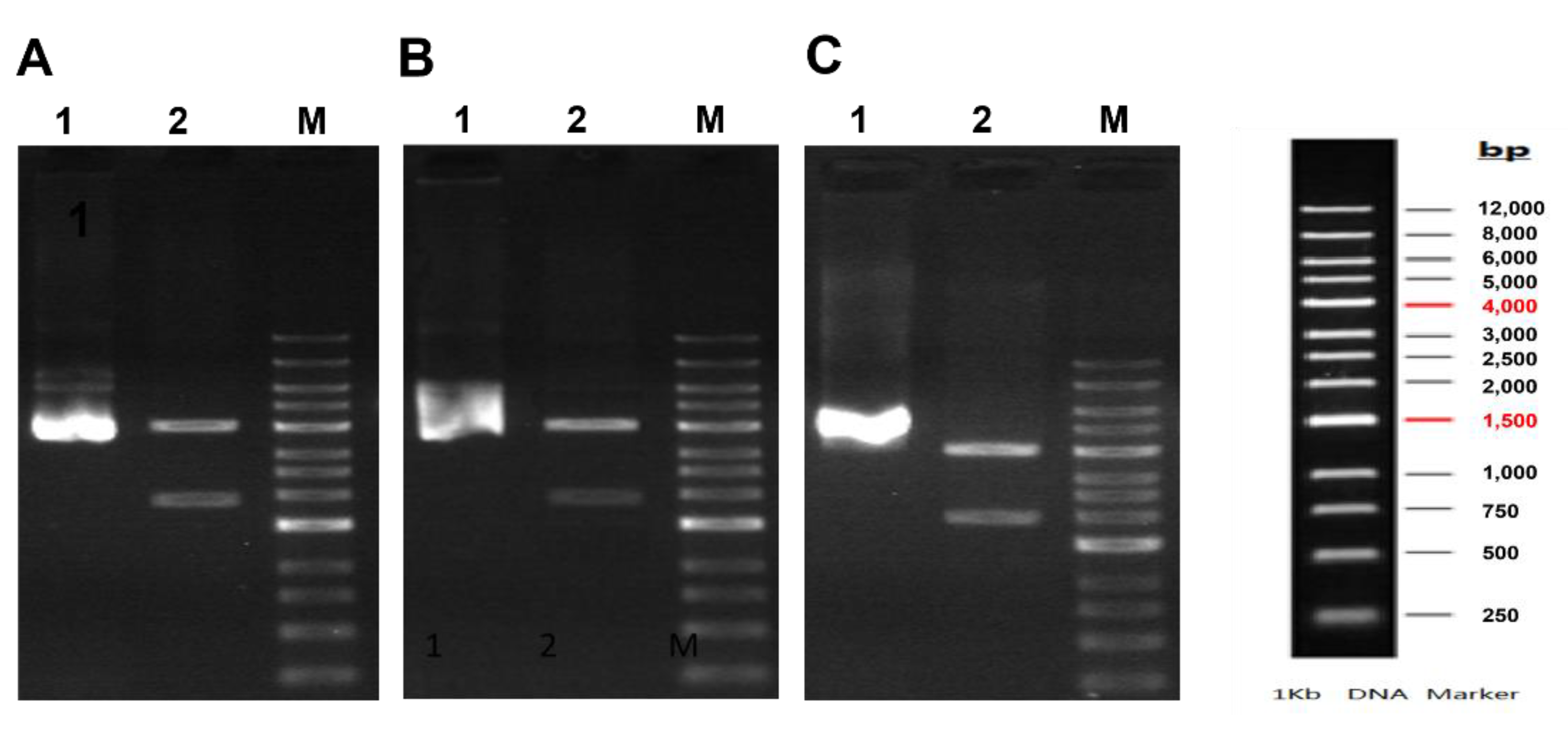
2.2. Chemosynthesis, Construction and Verification of Recombinant Plasmids with Pfk13-WT-Bac, Pfk13-F446I-Bac, and Pfk13-C580Y-Bac
2.3. Transfection and Collection of SF9 Cell with Pfk13-WT, Pfk13-F446I and Pfk13-C580Y
2.4. In Vitro Purification of Recombinant Protein of Pfk13-WT, Pfk13-F446I and Pfk13-C580Y
2.5. Western Blot
2.6. Protein Concentration Determination
2.7. Sequence Analysis
2.8. AlphaFold Modelling Analysis
2.9. Code Availability
3. Results
3.1. Constuction of Recombinant Pfk13-WT, Pfk13-F446I and Pfk13-C580Y
3.2. Expression of Pfk13-WT, Pfk13-F446I and Pfk13-C580Y in SF9 Cell
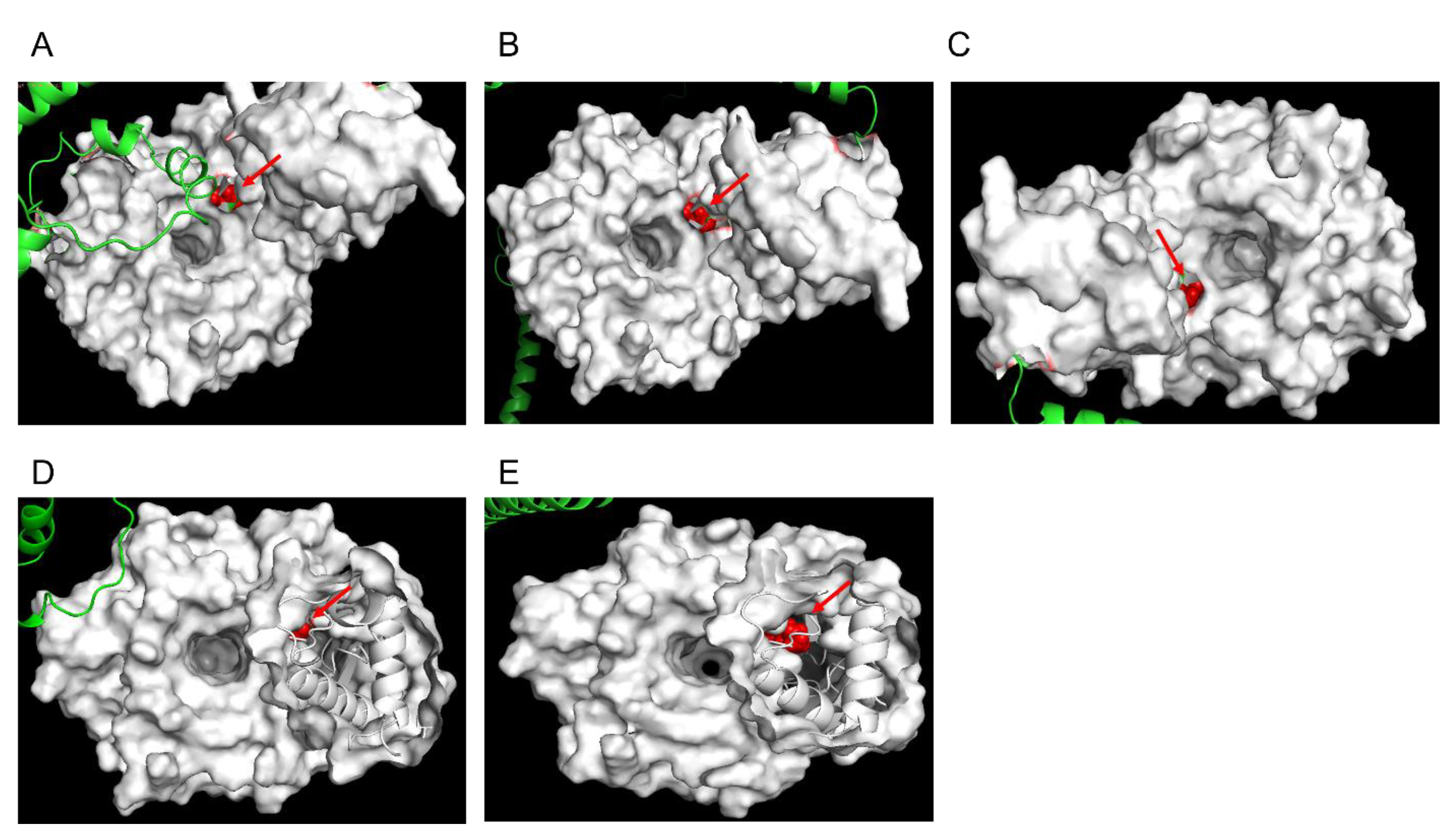
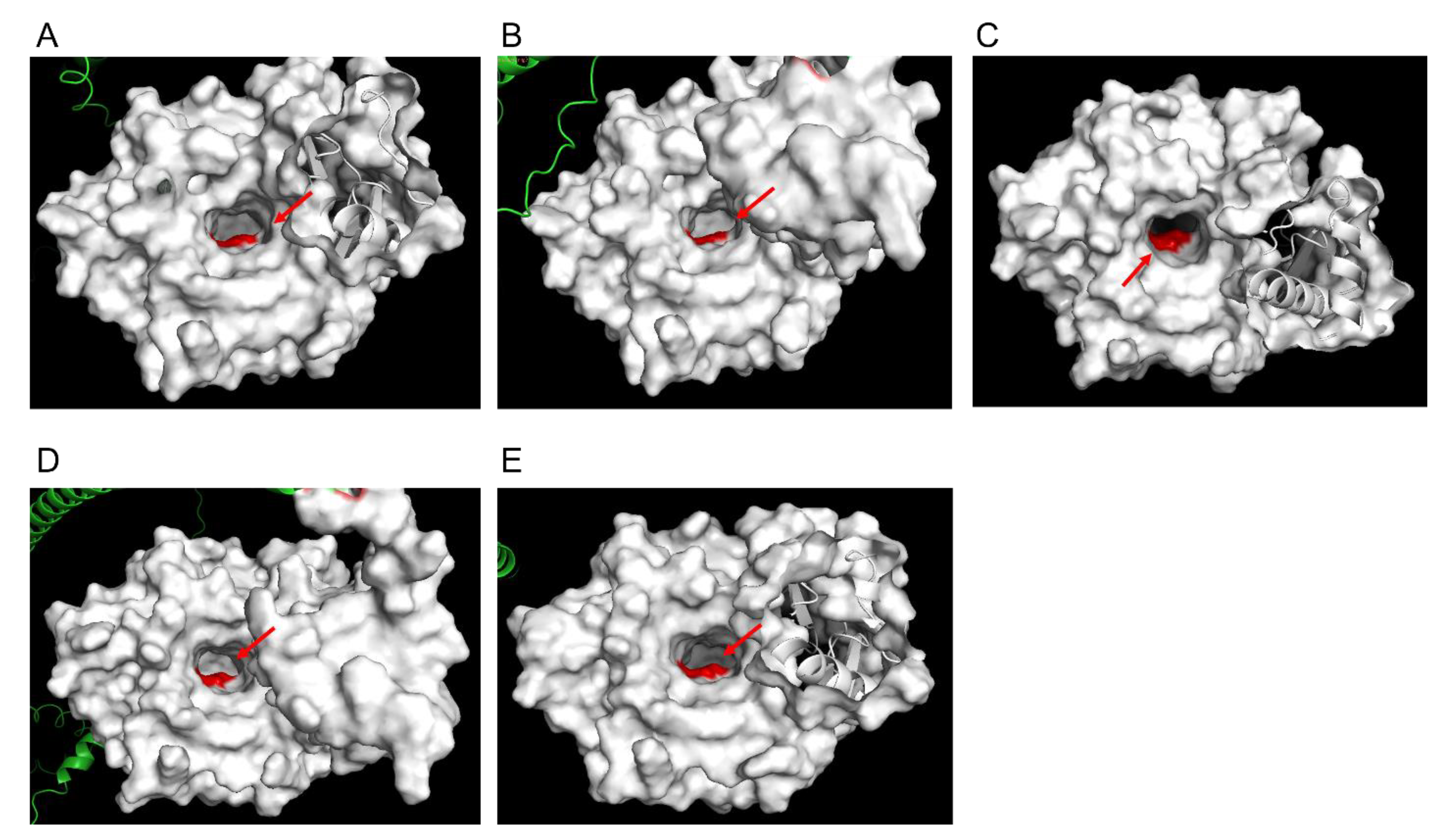
3.3. Structural Modelling of Pfk13-F446I and Pfk13-C580Y
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, X.-Z.; Wu, J. Zoonotic Transmission and Host Switches of Malaria Parasites. Zoonoses 2021, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Eastman, R.T.; Fidock, D.A. Artemisinin-based combination therapies: A vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 2009, 7, 864–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariey, F.; Witkowski, B.; Amaratunga, C.; Beghain, J.; Langlois, A.-C.; Khim, N.; Kim, S.; Duru, V.; Bouchier, C.; Ma, L.; et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 2014, 505, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of Artemisinin Resistance in Plasmodium falciparum Malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Witkowski, B.; Amaratunga, C.; Khim, N.; Sreng, S.; Chim, P.; Kim, S.; Lim, P.; Mao, S.; Sopha, C.; Sam, B.; et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In Vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013, 13, 1043–1049. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Mita, T.; Tachibana, S.-I.; Hashimoto, M.; Hirai, M. Plasmodium falciparum kelch 13: A potential molecular marker for tackling artemisinin-resistant malaria parasites. Expert Rev. Anti-Infect. Ther. 2015, 14, 125–135. [Google Scholar] [CrossRef]
- Fairhurst, R.M. Understanding artemisinin-resistant malaria: What a difference a year makes. Curr. Opin. Infect. Dis. 2015, 28, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Tilley, L.; Straimer, J.; Gnädig, N.F.; Ralph, S.A.; Fidock, D.A. Artemisinin Action and Resistance in Plasmodium falciparum. Trends Parasitol. 2016, 32, 682–696. [Google Scholar] [CrossRef] [Green Version]
- Phompradit, P.; Chaijaroenkul, W.; Na-Bangchang, K. Cellular mechanisms of action and resistance of Plasmodium falciparum to artemisinin. Parasitol. Res. 2017, 116, 3331–3339. [Google Scholar] [CrossRef]
- World Health Organization. World Malaria Report 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Tun, K.M.; Jeeyapant, A.; Imwong, M.; Thein, M.; Aung, S.S.M.; Hlaing, T.M.; Yuentrakul, P.; Promnarate, C.; Dhorda, M.; Woodrow, C.J.; et al. Parasite clearance rates in Upper Myanmar indicate a distinctive artemisinin resistance phenotype: A therapeutic efficacy study. Malar. J. 2016, 15, 185. [Google Scholar] [CrossRef]
- Zaw, M.T.; Lin, Z.; Emran, N.A. Importance of kelch 13 C580Y mutation in the studies of artemisinin resistance in Plasmodium falciparum in Greater Mekong Subregion. J. Microbiol. Immunol. Infect. 2019, 53, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Aninagyei, E.; Duedu, K.O.; Rufai, T.; Tetteh, C.D.; Chandi, M.G.; Ampomah, P.; Acheampong, D.O. Characterization of putative drug resistant biomarkers in Plasmodium falciparum isolated from Ghanaian blood donors. BMC Infect. Dis. 2020, 20, 533. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, L.C.; Cox, H.; Early, A.M.; Mok, S.; Lazrek, Y.; Paquet, J.-C.; Ade, M.-P.; Lucchi, N.; Grant, Q.; Udhayakumar, V.; et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife 2020, 9, e51015. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhou, D.; Lin, Y.; Xiao, H.; Yan, H.; Xia, Z. Amplification of pfmdr1, pfcrt, pvmdr1, and K13 Propeller Polymorphisms Associated with Plasmodium falciparum and Plasmodium vivax Isolates from the China-Myanmar Border. Antimicrob. Agents Chemother. 2015, 59, 2554–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Kong, X.; Xu, D.; Yan, H.; Zhou, H.; Tu, H.; Lin, K. Investigation and Evaluation of Genetic Diversity of Plasmodium falciparum Kelch 13 Polymorphisms Imported from Southeast Asia and Africa in Southern China. Front. Public Health 2019, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, Y.; Zhao, Y.; Ye, R.; Zhang, D.; Pan, W. Introduction of F446I mutation in the K13 propeller gene leads to increased ring survival rates in Plasmodium falciparum isolates. Malar. J. 2018, 17, 248. [Google Scholar] [CrossRef] [Green Version]
- Pazhayam, N.M.; Chhibber-Goel, J.; Sharma, A. New leads for drug repurposing against malaria. Drug Discov. Today 2018, 24, 263–271. [Google Scholar] [CrossRef]
- Singh, G.P.; Goel, P.; Sharma, A. Structural mapping of Kelch13 mutations associated with artemisinin resistance in malaria. J. Struct. Funct. Genom. 2016, 17, 51–56. [Google Scholar] [CrossRef]
- Haldar, K.; Bhattacharjee, S.; Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Sharma, A. Profiles of Kelch mutations in Plasmodium falciparum across South Asia and their implications for tracking drug resistance. Int. J. Parasitol. Drugs Drug Resist. 2019, 11, 49–58. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, L.; Xia, Z.G.; Xiao, N. Malaria elimination program in China: An eminent milestone in the anti-malaria campaign and challenges in the post-elimination stage. Chin. J. Parasitol. Parasit. Dis. 2021, 39, 421–428. [Google Scholar]
- Feng, J.; Zhang, L.; Xia, Z.G.; Zhou, S.S.; Xiao, N. Malaria-free certification in China: Achievements and lessons learned from the national malaria elimination programme. Zoonoses 2021, 1, 3–6. [Google Scholar] [CrossRef]
- Feng, J.; Li, J.; Yan, H.; Feng, X.; Xia, Z. Evaluation of Antimalarial Resistance Marker Polymorphism in Returned Migrant Workers in China. Antimicrob. Agents Chemother. 2015, 59, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Xu, D.; Kong, X.; Lin, K.; Yan, H.; Feng, X.; Tu, H.; Xia, Z. Characterization of pfmdr1, pfcrt, pfK13, pfubp1, and pfap2mu in Travelers Returning from Africa with Plasmodium falciparum Infections Reported in China from 2014 to 2018. Antimicrob. Agents Chemother. 2021, 65, e0271720. [Google Scholar] [CrossRef]
- Goel, N.; Dhiman, K.; Kalidas, N.; Mukhopadhyay, A.; Ashish, F.; Bhattacharjee, S. Plasmodium falciparum Kelch13 and its artemisinin-resistant mutants assemble as hexamers in solution: A SAXS data driven shape restoration study. FEBS J. 2022, 289, 4935–4962. [Google Scholar] [CrossRef]
- Lee, S.-M.; Hickey, J.M.; Miura, K.; Joshi, S.B.; Volkin, D.B.; King, C.R.; Plieskatt, J.L. A C-terminal Pfs48/45 malaria transmission-blocking vaccine candidate produced in the baculovirus expression system. Sci. Rep. 2020, 10, 395. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-M.; Wu, Y.; Hickey, J.M.; Miura, K.; Whitaker, N.; Joshi, S.B.; Volkin, D.B.; King, C.R.; Plieskatt, J. The Pfs230 N-terminal fragment, Pfs230D1+: Expression and characterization of a potential malaria transmission-blocking vaccine candidate. Malar. J. 2019, 18, 356. [Google Scholar] [CrossRef]
- Niu, G.; Cui, Y.; Wang, X.; Keleta, Y.; Li, J. Studies of the Parasite-Midgut Interaction Reveal Plasmodium Proteins Important for Malaria Transmission to Mosquitoes. Front. Cell Infect. Microbiol. 2021, 11, 654216. [Google Scholar] [CrossRef]
- Gibbons, J.; Button-Simons, K.A.; Adapa, S.R.; Li, S.; Pietsch, M.; Zhang, M.; Liao, X.; Adams, J.H.; Ferdig, M.T.; Jiang, R.H.Y. Altered expression of K13 disrupts DNA replication and repair in Plasmodium falciparum. BMC Genom. 2018, 19, 849. [Google Scholar] [CrossRef]
- Coppée, R.; Jeffares, D.C.; Miteva, M.A.; Sabbagh, A.; Clain, J. Comparative structural and evolutionary analyses predict functional sites in the artemisinin resistance malaria protein K13. Sci. Rep. 2019, 9, 10675. [Google Scholar] [CrossRef] [Green Version]
- Gnädig, N.F.; Stokes, B.H.; Edwards, R.L.; Kalantarov, G.F.; Heimsch, K.C.; Kuderjavy, M.; Crane, A.; Lee, M.C.S.; Straimer, J.; Becker, K.; et al. Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog. 2020, 16, e1008482. [Google Scholar] [CrossRef] [PubMed]
- Boizot, A.; Talmat-Amar, Y.; Morrogh, D.; Kuntz, N.L.; Halbert, C.; Chabrol, B.; Houlden, H.; Stojkovic, T.; Schulman, B.A.; Rautenstrauss, B.; et al. The instability of the BTB-KELCH protein Gigaxonin causes Giant Axonal Neuropathy and constitutes a new penetrant and specific diagnostic test. Acta Neuropathol. Commun. 2014, 2, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, L.; Balasco, N.; Smaldone, G.; Berisio, R.; Ruggiero, A.; Vitagliano, L. AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family. Biomolecules 2021, 11, 1862. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.J.; Courtney, J.M.; Shen, Y.; Ying, J.; Bax, A. Concordance of X-ray and AlphaFold2 Models of SARS-CoV-2 Main Protease with Residual Dipolar Couplings Measured in Solution. J. Am. Chem. Soc. 2021, 143, 19306–19310. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Taldaev, A.; Lisitsa, A.V.; Ponomarenko, E.A.; Archakov, A.I. Prediction of Monomeric and Dimeric Structures of CYP102A1 Using AlphaFold2 and AlphaFold Multimer and Assessment of Point Mutation Effect on the Efficiency of Intra- and Interprotein Electron Transfer. Molecules 2022, 27, 1386. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Thornton, J.M. PDBsum extras: SARS-CoV -2 and AlphaFold models. Protein Sci. 2021, 31, 283–289. [Google Scholar] [CrossRef]
- Torrado, R.P.; Yamada, D.; Defossez, P.-A. Born to bind: The BTB protein–protein interaction domain. BioEssays 2006, 28, 1194–1202. [Google Scholar] [CrossRef]
- Furukawa, M.; He, Y.J.; Borchers, C.; Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 2003, 5, 1001–1007. [Google Scholar] [CrossRef]
- Maekawa, M.; Higashiyama, S. The Roles of SPOP in DNA Damage Response and DNA Replication. Int. J. Mol. Sci. 2020, 21, 7293. [Google Scholar] [CrossRef]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.S.; Nandra, S.K.; Privé, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef]

| Model Type | Elements | x | Y | z |
|---|---|---|---|---|
| WT (PDB 4yy8) | N | −17.97 | −26.264 | 18.84 |
| WT (PDB 4yy8) | C | −18.141 | −24.822 | 18.834 |
| WT (PDB 4yy8) | C | −19.235 | −24.407 | 17.863 |
| WT (PDB 4yy8) | O | −19.36 | −24.961 | 16.775 |
| WT (PDB 4yy8) 1 | H | −0.39 | −3.488 | −1.942 |
| Model C580Y1 | N | −0.003 | −4.254 | −1.409 |
| Model C580Y1 | C | −0.698 | −4.612 | −0.171 |
| Model C580Y1 | C | −2.213 | −4.631 | −0.346 |
| Model C580Y1 | O | −2.805 | −3.605 | −0.643 |
| Model C580Y2 | N | −1.574 | 5.829 | −8.52 |
| Model C580Y2 | C | −2.887 | 5.914 | −7.874 |
| Model C580Y2 | C | −2.776 | 6.238 | −6.389 |
| Model C580Y2 | O | −2.229 | 5.443 | −5.637 |
| Model C580Y3 | N | 4.815 | −7.399 | −0.246 |
| Model C580Y3 | C | 3.858 | −8.028 | 0.66 |
| Model C580Y3 | C | 2.45 | −7.528 | 0.367 |
| Model C580Y3 | O | 2.2 | −6.335 | 0.442 |
| Model C580Y4 | N | −2.534 | 5.28 | −2.05 |
| Model C580Y4 | C | −3.805 | 5.291 | −1.329 |
| Model C580Y4 | C | −3.581 | 4.962 | 0.139 |
| Model C580Y4 | O | −3.097 | 3.886 | 0.458 |
| Model C580Y5 | N | 5.558 | −3.18 | 3.363 |
| Model C580Y5 | C | 4.469 | −3.103 | 4.333 |
| Model C580Y5 | C | 3.169 | −3.571 | 3.697 |
| Model C580Y5 | O | 2.703 | −2.969 | 2.743 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Feng, J.; Chen, M. Structural Modelling Prediction of Recombinant Plasmodium falciparum K13-F446I and K13-C580Y Gene by AlphaFold Method and Heterologous Expression in Spodoptera frugiperda 9 Cells. Pathogens 2022, 11, 1271. https://doi.org/10.3390/pathogens11111271
Yan H, Feng J, Chen M. Structural Modelling Prediction of Recombinant Plasmodium falciparum K13-F446I and K13-C580Y Gene by AlphaFold Method and Heterologous Expression in Spodoptera frugiperda 9 Cells. Pathogens. 2022; 11(11):1271. https://doi.org/10.3390/pathogens11111271
Chicago/Turabian StyleYan, He, Jun Feng, and Min Chen. 2022. "Structural Modelling Prediction of Recombinant Plasmodium falciparum K13-F446I and K13-C580Y Gene by AlphaFold Method and Heterologous Expression in Spodoptera frugiperda 9 Cells" Pathogens 11, no. 11: 1271. https://doi.org/10.3390/pathogens11111271






