Potential Pathogenicity of Aeromonas spp. Recovered in River Water, Soil, and Vegetation from a Natural Recreational Area
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection and Processing
2.2. Aeromonas Quantification by Plate Counting
2.3. Aeromonas Quantification by qPCR
2.4. Aeromonas Quantification by Most Probable Number (MPN)
2.5. Bacterial Strains Maintenance and Culture Conditions
2.6. DNA Extraction and Genus-Level Identification Based on GCAT Gene
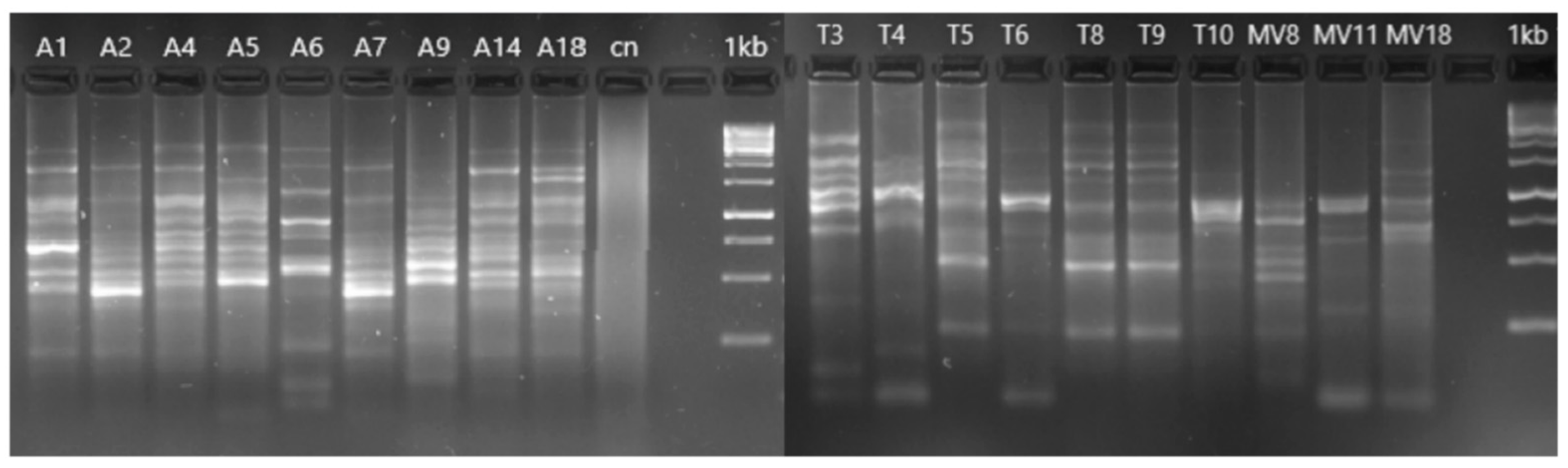
2.7. Genotyping of Aeromonas Strains
2.8. Identification of the Aeromonas Species Based on the rpoD Gene
2.9. Detection of Virulence Genes
2.10. Macrophages Growth Conditions and Infection
2.11. Analysis of the Expression of ascF-G and ast Genes after Infecting Macrophages
2.12. Quantification of Cell Damage in Macrophages
2.13. Intracellular Bacterial Survival of Aeromonas Strains in Macrophages
2.14. Antimicrobial Susceptibility Profile
2.15. Statistical Analysis
3. Results
3.1. Aeromonas Quantification
3.2. Genus-Level Identification Based on the GCAT Gene
3.3. Genotyping of Aeromonas Isolates
3.4. Species-Level Identification Based on the rpoD Gene
3.5. Identification of Virulence Genes
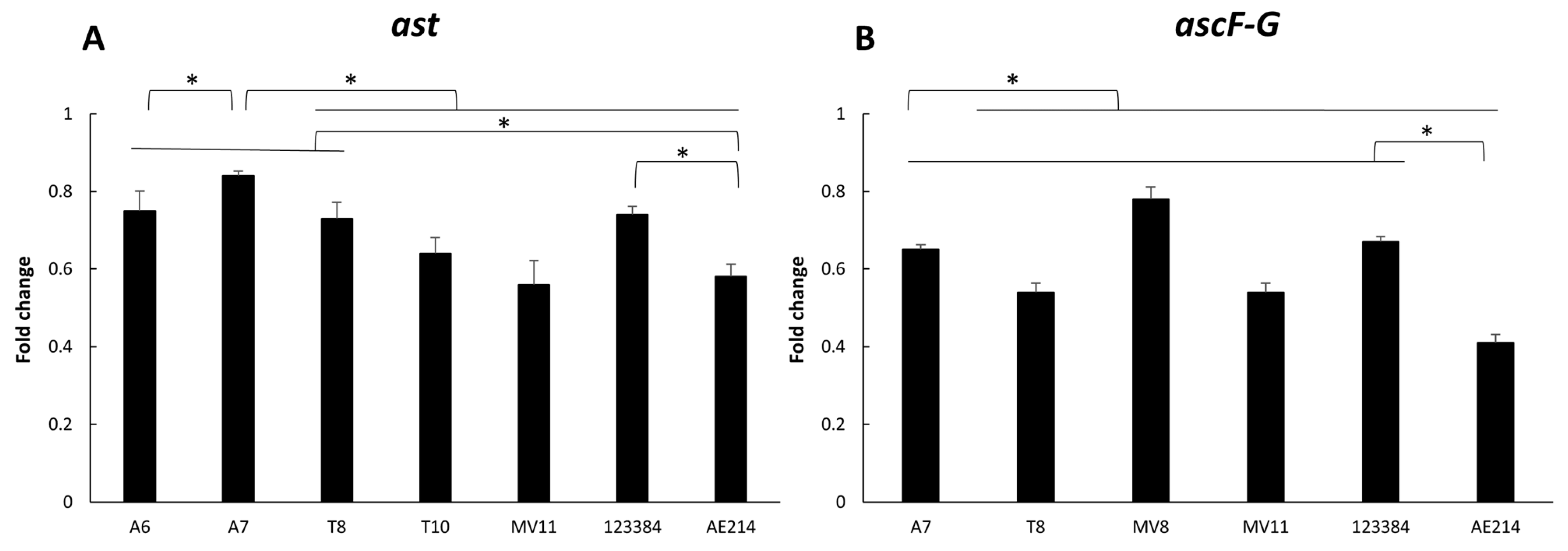
3.6. Virulence-Associated Gene Expression
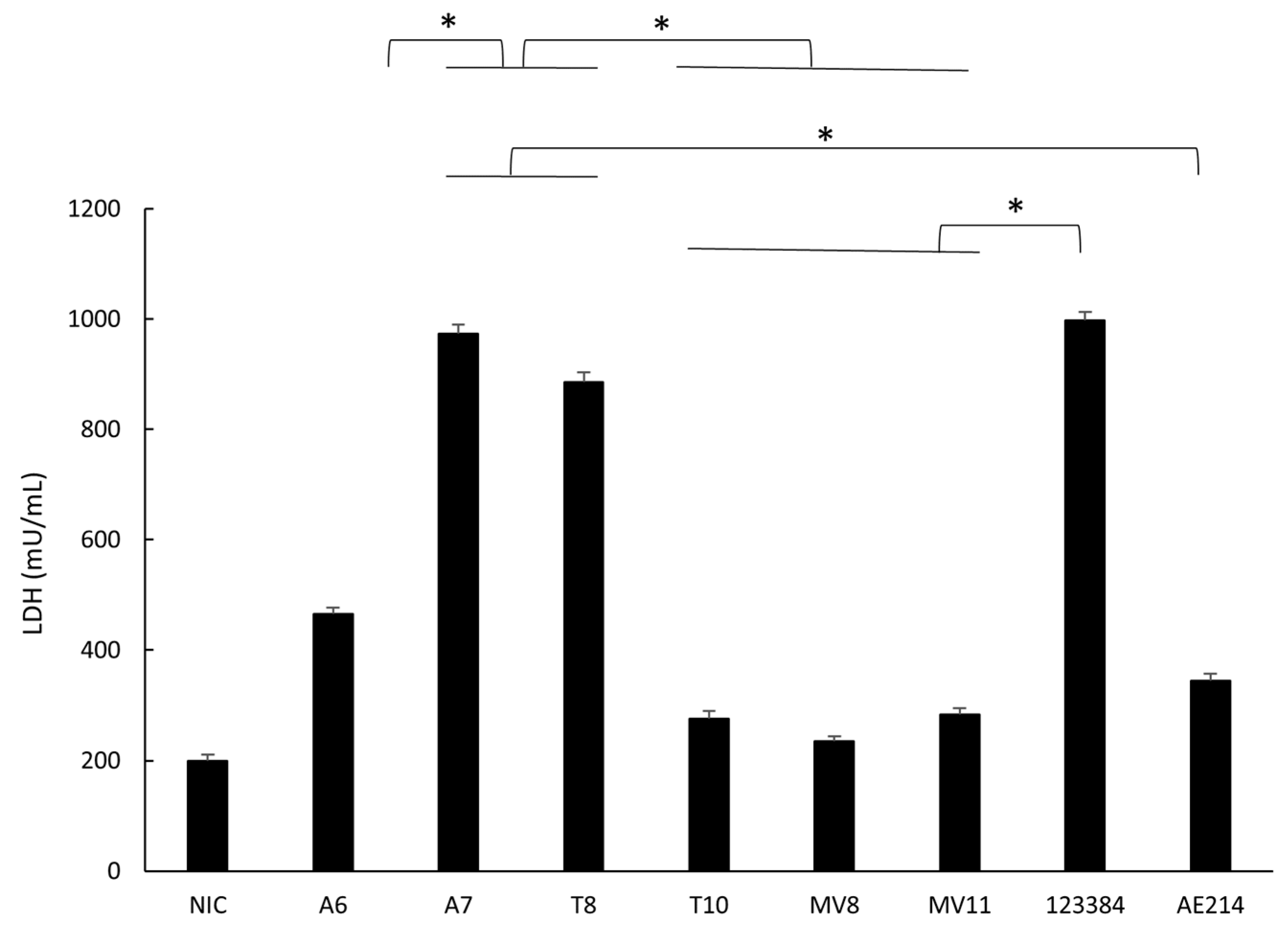
3.7. Quantification of Cell Damage in Macrophages
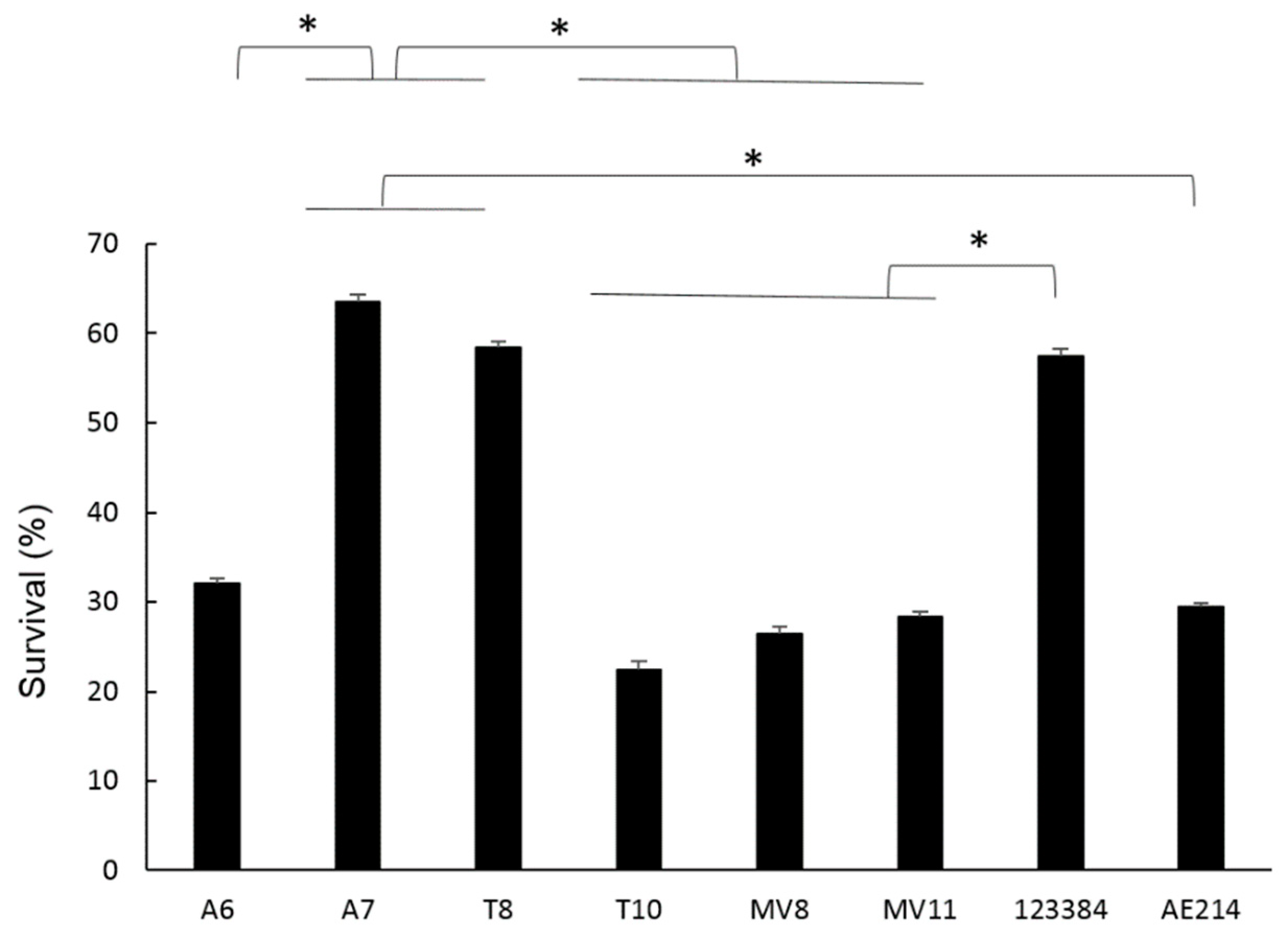
3.8. Intracellular Bacterial Survival of Aeromonas Strains
3.9. Antimicrobial Susceptibility Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Araujo, R.M.; Pares, R.; Lucena, F. The effect of terrestrial effluents on the incidence of Aeromonas spp. in coastal waters. J. Appl. Bacteriol. 1990, 69, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, C.M.; Coote, B.G.; Ashbolt, N.J.; Stevenson, I.M. Relationships between indicators, pathogens and water quality in an estuarine system. Water Res. 1996, 30, 2045–2054. [Google Scholar] [CrossRef]
- Borrell, N.; Figueras, M.J.; Guarro, J. Phenotypic identification of Aeromonas genomospecies from clinical and environmental sources. Can. J. Microbiol. 1998, 44, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Suarez-Franquet, A.; Chacón, M.R.; Soler, L.; Navarro, M.; Alejandre, C.; Grasa, B.; Martínez-Murcia, A.J.; Guarro, J. First record of the rare species Aeromonas culicicola from a drinking water supply. Appl. Environ. Microbiol. 2005, 71, 538–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLellan, S.L.; Huse, S.M.; Mueller-Spitz, S.R.; Andreishcheva, E.N.; Sogin, M.L. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 2010, 12, 378–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pablos, M.; Remacha, M.A.; Rodríguez-Calleja, J.M.; Santos, J.A.; Otero, A.; García-López, M.L. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Aravena-Román, M.; Beaz-Hidalgo, R.; Inglis, T.J.J.; Riley, T.V.; Martínez-Murcia, A.J.; Chang, B.J.; Figueras, M.J. Aeromonas australiensis sp. nov., isolated from irrigation water. Int. J. Syst. Evol. Microbiol. 2013, 63, 2270–2276. [Google Scholar] [CrossRef] [Green Version]
- Latif-Eugenín, F.; Beaz-Hidalgo, R.; Silvera-Simón, C.; Fernandez-Cassi, X.; Figueras, M.J. Chlorinated and ultraviolet radiation -treated reclaimed irrigation water is the source of Aeromonas found in vegetables used for human consumption. Environ. Res. 2017, 154, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Figueras Salvat, M.J.; Ashbolt, N. Aeromonas. Water Sanit. 21st Century Health Microbiol. Asp. Excreta Wastewater Manag. Glob. Water Pathog. Proj. 2019. [Google Scholar] [CrossRef] [Green Version]
- Rusiñol, M.; Hundesa, A.; Cárdenas-Youngs, Y.; Fernández-Bravo, A.; Pérez-Cataluña, A.; Moreno-Mesonero, L.; Moreno, Y.; Calvo, M.; Alonso, J.L.; Figueras, M.J.; et al. Microbiological contamination of conventional and reclaimed irrigation water: Evaluation and management measures. Sci. Total Environ. 2020, 710, 136298. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, S.W.; Carnahan, A.M.; Brayton, P.R.; Fanning, G.R.; Almazan, R.; Drabick, C.; Trudo, E.W.; Colwell, R.R. Aeromonas jandaei and Aeromonas veronii dual infection of a human wound following aquatic exposure. J. Clin. Microbiol. 1991, 29, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, J.R.; Brum, G.; Fernandes, A.; Biscaia, I.; Salvado Correia, M.J.; Bastardo, J. Aeromonas hydrophila fulminant pneumonia in a fit young man. Thorax 1992, 47, 482–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vally, H.; Whittle, A.; Cameron, S.; Dowse, G.K.; Watson, T. Outbreak of Aeromonas hydrophila wound infections associated with mud football. Clin. Infect. Dis. 2004, 38, 1084–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimbrough, R.C.; Winn, R.E.; Jeter, R.M.; Warren, W.J.; Huddleston, J.R.; Zak, J.C. Aeromonas infection from river and playa lake waters in west texas and southeastern New Mexico. Southwest Respir. Crit. Care Chronicles 2016, 4, 19. [Google Scholar] [CrossRef] [Green Version]
- Grim, C.J.; Kozlova, E.V.; Ponnusamy, D.; Fitts, E.C.; Sha, J.; Kirtley, M.L.; van Lier, C.J.; Tiner, B.L.; Erova, T.E.; Joseph, S.J.; et al. Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila. Appl. Environ. Microbiol. 2014, 80, 4162–4183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnusamy, D.; Kozlova, E.V.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci. USA. 2016, 113, 722–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueras, M.J.; Beaz-Hidalgo, R. Aeromonas infections in humans. In Aeromonas; Academic Press: Norfolk, UK, 2015; pp. 65–108. [Google Scholar]
- Pianetti, A.; Sabatini, L.; Bruscolini, F.; Chiaverini, F.; Cecchetti, G. Faecal contamination indicators, Salmonella, Vibrio and Aeromonas in water used for the irrigation of agricultural products. Epidemiol. Infect. 2004, 132, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Al-Jassim, N.; Ansari, M.I.; Harb, M.; Hong, P.Y. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: Is the treated wastewater safe to reuse for agricultural irrigation? Water Res. 2015, 73, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Tomás, J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012, 2012, 256261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Figueira, V.; Vaz-Moreira, I.; Silva, M.; Manaia, C.M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 2011, 45, 5599–5611. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Shih, D.Y.C.; Wang, J.Y.; Yang, S.S. Molecular characterization of class 1 integrons and antimicrobial resistance in Aeromonas strains from foodborne outbreak-suspect samples and environmental sources in Taiwan. Diagn. Microbiol. Infect. Dis. 2007, 59, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; von Czapiewski, E.; Kaspar, H.; Wallmann, J.; Michael, G.B.; Steinacker, U.; Schwarz, S. Molecular basis of sulfonamide and trimethoprim resistance in fish-pathogenic Aeromonas isolates. Appl. Environ. Microbiol. 2011, 77, 7147–7150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, A.; Henriques, I.; Ribeiro, R.; Correia, A. Prevalence and Characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 2007, 60, 1243–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, M.J.; Martínez-Murcia, A.; Esteves, A.C.; Correia, A.; Saavedra, M.J. Phylogenetic Diversity, antibiotic resistance and virulence traits of Aeromonas spp. from untreated waters for human consumption. Int. J. Food Microbiol. 2012, 159, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Chacón, M.R.; Castro-Escarpulli, G.; Soler, L.; Guarro, J.; Figueras, M.J. A DNA probe specific for Aeromonas colonies. Diagn. Microbiol. Infect. Dis. 2002, 44, 221–225. [Google Scholar] [CrossRef]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive dna sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Soler, L.; Yáñez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalán, V.; Figueras, M.J.; Martínez-Murcia, A.J. Phylogenetic Analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519. [Google Scholar] [CrossRef]
- Merino, S.; Gavín, R.; Vilches, S.; Shaw, J.G.; Tomás, J.M. A Colonization factor (production of lateral flagella) of mesophilic Aeromonas spp. is inactive in Aeromonas salmonicida strains. Appl. Environ. Microbiol. 2003, 69, 663–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, A.K.; Peterson, J.W.; Xu, X.J.; Coppenhaver, D.H.; Houston, C.W. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 1996, 21, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hoel, S.; Lunestad, B.T.; Lerfall, J.; Jakobsen, A.N. Aeromonas spp. Isolated from ready-to-eat seafood on the norwegian market: Prevalence, putative virulence factors and antimicrobial resistance. J. Appl. Microbiol. 2021, 130, 1380–1393. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Clark, C.G.; Rodgerst, F.G. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 shiga toxin family by multiplex PCR. J. Clin. Microbiol. 2002, 40, 3613–3619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chacón, M.R.; Soler, L.; Groisman, E.A.; Guarro, J.; Figueras, M.J. Type III Secretion System Genes in Clinical Aeromonas Isolates. J. Clin. Microbiol. 2004, 42, 1285–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houf, K.; De Zutter, L.; Van Hoof, J.; Vandamme, P. Assessment of the genetic diversity among Arcobacters isolated from poultry products by using two PCR-based typing methods. Appl. Environ. Microbiol. 2002, 68, 2172–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Rama, D.; Esendagli, G.; Guc, D. Expression of chemokine-like receptor 1 (CMKLR1) on J744A.1 macrophages co-cultured with fibroblast and/or tumor cells: Modeling the influence of microenvironment. Cell. Immunol. 2011, 271, 134–140. [Google Scholar] [CrossRef]
- Murciano, C.; Hor, L.I.; Amaro, C. Host-pathogen interactions in Vibrio vulnificus: Responses of monocytes and vascular endothelial cells to live bacteria. Future Microbiol. 2015, 10, 471–487. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. Immune response of the monocytic cell line THP-1 against six Aeromonas spp. Front. Immunol. 2022, 13, 875689. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bravo, A.; López-Fernández, L.; Figueras, M.J. The metallochaperone encoding gene HypA Is widely distributed among pathogenic Aeromonas spp. and its expression is increased under acidic pH and within macrophages. Microorganisms 2019, 7, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, M.A.; Burke, V.; Chang, B.J. Invasion of HEp-2 cells by fecal isolates of Aeromonas hydrophila. Infect. Immun. 1985, 47, 680–683. [Google Scholar] [CrossRef] [Green Version]
- CLSI Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2020. [Google Scholar]
- Mohanty, S.; Ali, S.M.; Singh, P.K. Necrotizing fasciitis and gas gangrene due to Aeromonas hydrophila in an immunocompetent host: A rare entity. IDCases 2022, 28, e01508. [Google Scholar] [CrossRef]
- Hasan, O.; Khan, W.; Jessar, M.; Pathan, A.Z.; Lakdawala, R.H. Bone graft donor site infection with a rare organism, Aeromonas hydrophila. a typical location, presentation and organism with 2 years follow-up. case report. Int. J. Surg. Case Rep. 2018, 51, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bravo, A.; Kilgore, P.B.; Andersson, J.A.; Blears, E.; Figueras, M.J.; Hasan, N.A.; Colwell, R.R.; Sha, J.; Chopra, A.K. T6SS and ExoA of flesh-eating Aeromonas hydrophila in peritonitis and necrotizing fasciitis during mono- and polymicrobial infections. Proc. Natl. Acad. Sci. USA 2019, 116, 24084–24092. [Google Scholar] [CrossRef]
- Botes, M.; De Kwaadsteniet, M.; Cloete, T.E. Application of quantitative PCR for the detection of microorganisms in water. Anal. Bioanal. Chem. 2013, 405, 91–108. [Google Scholar] [CrossRef]
- Loozen, G.; Boon, N.; Pauwels, M.; Quirynen, M.; Teughels, W. Live/dead real-time polymerase chain reaction to assess new therapies against dental plaque-related pathologies. Mol. Oral Microbiol. 2011, 26, 253–261. [Google Scholar] [CrossRef]
- Latif-Eugenín, F.; Beaz-Hidalgo, R.; Figueras, M.J. Evaluation of different conditions and culture media for the recovery of Aeromonas spp. from water and shellfish samples. J. Appl. Microbiol. 2016, 121, 883–891. [Google Scholar] [CrossRef]
- Latif-Eugenín, F.; Beaz-Hidalgo, R.; Figueras, M.J. A Culture Independent method for the detection of Aeromonas sp. from water samples. Ital. J. food Saf. 2016, 5, 11–14. [Google Scholar] [CrossRef]
- Figueras, M.J.; Beaz Hidalgo, R.; Collado, L.; Martínez-Murcias, A. Recommendations for a new bacterial species description based on analyses of the unrelated genera Aeromonas and Arcobacter. Bull. BISMiS 2011, 2, 1–16. [Google Scholar]
- Gavín, R.; Rabaan, A.A.; Merino, S.; Tomás, J.M.; Gryllos, I.; Shaw, J.G. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 2002, 43, 383–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, P.G.; Santos, P.A.; Bello, A.R.; Freitas-Almeida, A.C. Association of Aeromonas caviae polar and lateral flagella with biofilm formation. Lett. Appl. Microbiol. 2011, 52, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Aravena-Román, M.; Inglis, T.J.J.; Riley, T.V.; Chang, B.J. Distribution of 13 virulence genes among clinical and environmental Aeromonas spp. in western australia. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1889–1895. [Google Scholar] [CrossRef]
- Alperi, A.; Figueras, M.J. Human isolates of Aeromonas possess shiga toxin genes (stx1 and stx2) highly similar to the most virulent gene variants of Escherichia coli. Clin. Microbiol. Infect. 2010, 16, 1563–1567. [Google Scholar] [CrossRef] [Green Version]
- Palma-Martínez, I.; Guerrero-Mandujano, A.; Ruiz-Ruiz, M.J.; Hernández-Cortez, C.; Molina-López, J.; Bocanegra-García, V.; Castro-Escarpulli, G. Active shiga-like toxin produced by some Aeromonas spp., isolated in mexico city. Front. Microbiol. 2016, 7, 1522. [Google Scholar] [CrossRef] [Green Version]
- Vilches, S.; Urgell, C.; Merino, S.; Chacón, M.R.; Soler, L.; Castro-Escarpulli, G.; Figueras, M.J.; Tomás, J.M. Complete type III secretion system of a mesophilic Aeromonas hydrophila strain. Appl. Environ. Microbiol. 2004, 70, 6914–6919. [Google Scholar] [CrossRef] [Green Version]
- Frey, J.; Origgi, F.C. Type III secretion system of Aeromonas salmonicida undermining the host’s immune response. Front. Mar. Sci. 2016, 3, 130. [Google Scholar] [CrossRef] [Green Version]
- Tseng, T.T.; Tyler, B.M.; Setubal, J.C. Protein secretion systems in bacterial-host associations, and their description in the gene ontology. BMC Microbiol. 2009, 9, S2. [Google Scholar] [CrossRef] [Green Version]
- Rangel, L.T.; Marden, J.; Colston, S.; Setubal, J.C.; Graf, J.; Gogarten, J.P. Identification and characterization of putative Aeromonas spp. t3ss effectors. PLoS ONE 2019, 14, e0214035. [Google Scholar] [CrossRef] [Green Version]
- Madara, J.L.; Stafford, J. Interferon-gamma directly affects barrier function of cultured intestinal epithelial monolayers. J. Clin. Investig. 1989, 83, 724–727. [Google Scholar] [CrossRef]
- Epple, H.J.; Mankertz, J.; Ignatius, R.; Liesenfeld, O.; Fromm, M.; Zeitz, M.; Chakraborty, T.; Schulzke, J.D. Aeromonas hydrophila beta-hemolysin induces active chloride secretion in colon epithelial cells (HT-29/B6). Infect. Immun. 2004, 72, 4848–4858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couto, C.R.A.; Oliveira, S.S.; Queiroz, M.L.P.; Freitas-Almeida, A.C. Interactions of clinical and environmental Aeromonas isolates with Caco-2 and HT29 intestinal epithelial cells. Lett. Appl. Microbiol. 2007, 45, 405–410. [Google Scholar] [CrossRef]
- Dos Santos, P.A.; Pereira, A.C.M.; Ferreira, A.F.; de Mattos Alves, M.A.; Rosa, A.C.P.; Freitas-Almeida, A.C. Adhesion, invasion, intracellular survival and cytotoxic activity of strains of Aeromonas spp. in HEp-2, Caco-2 and T-84 cell lines. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2015, 107, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Ribeiro, M.; Correia-Branco, A.; Domínguez-Perles, R.; Martel, F.; Saavedra, M.J.; Simões, M. Virulence, attachment and invasion of Caco-2 cells by multidrug-resistant bacteria isolated from wild animals. Microb. Pathog. 2019, 128, 230–235. [Google Scholar] [CrossRef] [PubMed]
- EUCAST Expert Rules Version 3.2. 20 February 2020. Available online: https://www.eucast.org/expert_rules_and_expected_phenotypes (accessed on 5 October 2022).
- Lamy, B.; Kodjo, A.; Laurent, F. Prospective nationwide study of Aeromonas infections in France. J. Clin. Microbiol. 2009, 47, 1234–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndi, O.L.; Barton, M.D. Incidence of class 1 integron and other antibiotic resistance determinants in Aeromonas spp. from rainbow trout farms in Australia. J. Fish Dis. 2011, 34, 589–599. [Google Scholar] [CrossRef]
- Aravena-Román, M.; Inglis, T.J.J.; Henderson, B.; Riley, T.V.; Chang, B.J. Antimicrobial susceptibilities of Aeromonas strains isolated from clinical and environmental sources to 26 antimicrobial agents. Antimicrob. Agents Chemother. 2012, 56, 1110–1112. [Google Scholar] [CrossRef] [Green Version]
- Scarano, C.; Piras, F.; Virdis, S.; Ziino, G.; Nuvoloni, R.; Dalmasso, A.; De Santis, E.P.L.; Spanu, C. Antibiotic resistance of Aeromonas ssp. strains isolated from Sparus aurata reared in italian mariculture farms. Int. J. Food Microbiol. 2018, 284, 91–97. [Google Scholar] [CrossRef]
- Carnahan, A.M.; Chakraborty, T.; Fanning, G.R.; Verma, D.; Ali, A.; Janda, J.M.; Joseph, S.W. Aeromonas trota sp. nov., an ampicillin-susceptible species isolated from clinical specimens. J. Clin. Microbiol. 1991, 29, 1206–1210. [Google Scholar] [CrossRef] [Green Version]
- Overman, T.L.; Janda, J.M. Antimicrobial susceptibility patterns of Aeromonas jandaei, A. schubertii, A. trota, and A. veronii biotype veronii. J. Clin. Microbiol. 1999, 37, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Denys, R.; Swings, J. DNA-DNA reassociation and phenotypic data indicate synonymy between Aeromonas enteropelogenes Schubert et al. 1990 and Aeromonas trota Carnahan et al. 1991. Int. J. Syst. Evol. Microbiol. 2002, 52, 1969–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbott, S.L.; Cheung, W.K.W.; Janda, J.M. The genus Aeromonas: Biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 2003, 41, 2348–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.C.; Ding, L.W.; Hsueh, P.R. Wound infection and septic shock due to Aeromonas trota in a patient with liver cirrhosis. Clin. Infect. Dis. 2007, 44, 1523–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallagassa, C.B.; Surek, M.; Vizzotto, B.S.; Prediger, K.C.; Moriel, B.; Wolf, S.; Weiss, V.; Cruz, L.M.; Assis, F.E.A.; Paludo, K.S.; et al. Characteristics of an Aeromonas trota strain isolated from cerebrospinal fluid. Microb. Pathog. 2018, 116, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.Y.; Jung, D.S.; Peck, K.R. Clinical and therapeutic implications of Aeromonas bacteremia: 14 years nation-wide experiences in Korea. Infect. Chemother. 2016, 48, 274–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Fan, Y.; Jiang, N.; Liu, W.; Shi, Y.; Zhao, J.; Zeng, L. Molecular characteristics and virulence analysis of eight Aeromonas hydrophila isolates obtained from diseased amur sturgeon acipenser schrenckii brandt, 1869. J. Vet. Med. Sci. 2018, 80, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.L.; Ko, W.C.; Wu, C.J. Complexity of β-lactamases among clinical Aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 2012, 45, 398–403. [Google Scholar] [CrossRef] [Green Version]
- Piotrowska, M.; Przygodzinska, D.; Matyjewicz, K.; Popowska, M. Occurrence and variety of β lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelo-Branco, D.d.S.C.M.; Guedes, G.M.d.M.; Brilhante, R.S.N.; Rocha, M.F.G.; Sidrim, J.J.C.; Moreira, J.L.B.; Cordeiro, R.d.A.; Sales, J.A.; Riello, G.B.; De Alencar, L.P.; et al. Virulence and antimicrobial susceptibility of clinical and environmental strains of Aeromonas spp. from northeastern Brazil. Can. J. Microbiol. 2015, 61, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Dhanapala, P.M.; Kalupahana, R.S.; Kalupahana, A.W.; Wijesekera, D.P.H.; Kottawatta, S.A.; Jayasekera, N.K.; Silva-Fletcher, A.; Jagoda, S.S.S.S. Characterization and antimicrobial resistance of environmental and clinical Aeromonas species isolated from fresh water ornamental fish and associated farming environment in Sri Lanka. Microorganisms 2021, 9, 2106. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence 5′-3′ | Target | Reference |
|---|---|---|---|
| GCAT-F | CTCCTGGAATCCCAAGTATCAG | GCAT | [29] |
| GCAT-R | GGCAGGTTGAACAGCAGTATCT | ||
| ERIC 1R | ATGTAAGCTCCTGGGGATTCAC | Genome | [30] |
| ERIC 2 | AAGTAAGTGACTGGGGTGAGCG | ||
| RpoD-Fs | GTCAATTCCGCCTGATGC | rpoD | [31] |
| RpoD-Rs | ATCATCTCGCGCATGTTGT | ||
| Laf1 | GGTCTGCGCATCCAACTC | lafA | [32] |
| Laf2 | GCTCCAGACGGTTGATG | ||
| alt-F | AAAGCGTCTGACAGCGAAGT | alt | [33] |
| alt-R | AGCGCATAGGCGTTCTCTT | ||
| ast-F | ATCGTCAGCGACAGCTCTT | ast | [34] |
| ast-R | CTCATCCCTTGGCTTGTTGT | ||
| Stx1-a | TCTCAGTGGGCGTTCTTATG | stx1 | [35] |
| Stx1-b | TACCCCCTCAACTGCTAATA | ||
| ascF-G-F | ATGAGGTCATCTGCTCGCGC | ascF-G | [36] |
| ascF-G-R | GGAGCACAACCATGGCTGAT |
| Gene | Product | Reference | A6 | A7 | T8 | T10 | MV8 | MV11 |
|---|---|---|---|---|---|---|---|---|
| laf | Lateral flagella structural protein | [32] | + | + | + | + | + | + |
| alt | Cytotonic enterotoxin | [33] | + | + | + | - | - | + |
| ast | Cytotonic enterotoxin | [34] | + | + | + | + | - | + |
| stx1 | Shiga toxin | [35] | - | - | - | - | - | - |
| ascF-G | T3SS structural protein | [36] | - | + | + | - | + | + |
| Antimicrobial (µg) | A6 | A7 | T8 | T10 | MV8 | MV11 |
|---|---|---|---|---|---|---|
| Ampicillin (10) | R | R | R | R | R | R |
| Cefuroxime (30) | I | S | I | R | R | R |
| Ceftriaxone (30) | S | S | S | I | R | R |
| Tetracycline (30) | S | I | I | I | I | I |
| Piperacillin/tazobactam (100/10) | S | S | R | I | I | R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerra, R.M.; Maleno, F.D.; Figueras, M.J.; Pujol-Bajador, I.; Fernández-Bravo, A. Potential Pathogenicity of Aeromonas spp. Recovered in River Water, Soil, and Vegetation from a Natural Recreational Area. Pathogens 2022, 11, 1382. https://doi.org/10.3390/pathogens11111382
Guerra RM, Maleno FD, Figueras MJ, Pujol-Bajador I, Fernández-Bravo A. Potential Pathogenicity of Aeromonas spp. Recovered in River Water, Soil, and Vegetation from a Natural Recreational Area. Pathogens. 2022; 11(11):1382. https://doi.org/10.3390/pathogens11111382
Chicago/Turabian StyleGuerra, Roberto M., Francisco Damián Maleno, Maria José Figueras, Isabel Pujol-Bajador, and Ana Fernández-Bravo. 2022. "Potential Pathogenicity of Aeromonas spp. Recovered in River Water, Soil, and Vegetation from a Natural Recreational Area" Pathogens 11, no. 11: 1382. https://doi.org/10.3390/pathogens11111382







