Whole Blood versus Plasma Samples—How Does the Type of Specimen Collected for Testing Affect the Monitoring of Cytomegalovirus Viremia?
Abstract
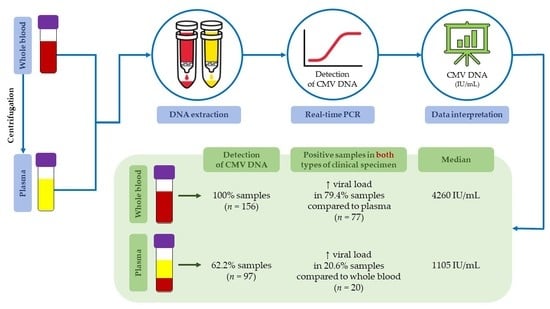
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. DNA Extraction
2.3. Nucleic Acid Amplification Test
2.4. Data Interpretation and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schottstedt, V.; Blümel, J.; Burger, R.; Drosten, C.; Gröner, A.; Gürtler, L.; Heiden, M.; Hildebrandt, M.; Jansen, B.; Montag-Lessing, T.; et al. Human Cytomegalovirus (HCMV)—Revised*. Transfus. Med. Hemother. 2010, 37, 365–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of Cytomegalovirus Seroprevalence and Demographic Characteristics Associated with Infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, J.L.; Schleiss, M.R. Prevention of Maternal Cytomegalovirus Infection: Current Status and Future Prospects. Int. J. Womens Health 2010, 2, 23–35. [Google Scholar] [PubMed] [Green Version]
- Dworsky, M.; Yow, M.; Stagno, S.; Pass, R.F.; Alford, C. Cytomegalovirus Infection of Breast Milk and Transmission in Infancy. Pediatrics 1983, 72, 295–299. [Google Scholar] [CrossRef]
- Stagno, S.; Reynolds, D.W.; Pass, R.F.; Alford, C.A. Breast Milk and the Risk of Cytomegalovirus Infection. N. Engl. J. Med. 1980, 302, 1073–1076. [Google Scholar] [CrossRef]
- Cannon, M.J.; Davis, K.F. Washing Our Hands of the Congenital Cytomegalovirus Disease Epidemic. BMC Public Health 2005, 5, 70. [Google Scholar] [CrossRef] [Green Version]
- CDC. About Cytomegalovirus and Congenital CMV Infection. Available online: https://www.cdc.gov/cmv/overview.html (accessed on 7 June 2022).
- Sinzger, C.; Digel, M.; Jahn, G. Cytomegalovirus Cell Tropism. In Human Cytomegalovirus; Shenk, T.E., Stinski, M.F., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 325, pp. 63–83. ISBN 978-3-540-77348-1. [Google Scholar]
- Britt, W. Manifestations of Human Cytomegalovirus Infection: Proposed Mechanisms of Acute and Chronic Disease. In Human Cytomegalovirus; Shenk, T.E., Stinski, M.F., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 325, pp. 417–470. ISBN 978-3-540-77348-1. [Google Scholar]
- Griffiths, P.; Reeves, M. Pathogenesis of Human Cytomegalovirus in the Immunocompromised Host. Nat. Rev. Microbiol. 2021, 19, 759–773. [Google Scholar] [CrossRef]
- Reeves, M.; Sinclair, J. Aspects of Human Cytomegalovirus Latency and Reactivation. In Human Cytomegalovirus; Shenk, T.E., Stinski, M.F., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2008; Volume 325, pp. 297–313. ISBN 978-3-540-77348-1. [Google Scholar]
- Taylor-Wiedeman, J.; Sissons, J.G.; Borysiewicz, L.K.; Sinclair, J.H. Monocytes Are a Major Site of Persistence of Human Cytomegalovirus in Peripheral Blood Mononuclear Cells. J. Gen. Virol. 1991, 72, 2059–2064. [Google Scholar] [CrossRef]
- Slobedman, B.; Mocarski, E.S. Quantitative Analysis of Latent Human Cytomegalovirus. J. Virol. 1999, 73, 4806–4812. [Google Scholar] [CrossRef] [Green Version]
- Slobedman, B.; Stern, J.L.; Cunningham, A.L.; Abendroth, A.; Abate, D.A.; Mocarski, E.S. Impact of Human Cytomegalovirus Latent Infection on Myeloid Progenitor Cell Gene Expression. J. Virol. 2004, 78, 4054–4062. [Google Scholar] [CrossRef]
- Griffiths, P.D.; Emery, V.C. Taming the Transplantation Troll by Targeting Terminase. N. Engl. J. Med. 2014, 370, 1844–1846. [Google Scholar] [CrossRef] [PubMed]
- Kotton, C.N.; Kumar, D.; Caliendo, A.M.; Huprikar, S.; Chou, S.; Danziger-Isakov, L.; Humar, A. The Transplantation Society International CMV Consensus Group the Third International Consensus Guidelines on the Management of Cytomegalovirus in Solid-Organ Transplantation. Transplantation 2018, 102, 900–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokorska-Śpiewak, M.; Niezgoda, A.; Gołkowska, M.; Czech-Kowalska, J.; Gruszfeld, D.; Dobrzańska, A.; Styczyński, J.; Marczyńska, M. Recommendations for the Diagnosis and Treatment of CMV Infections. Polish Society of Epidemiology and Infectious Diseases. Przegl. Epidemiol. 2016, 70, 297–310. [Google Scholar] [PubMed]
- Teira, P.; Battiwalla, M.; Ramanathan, M.; Barrett, A.J.; Ahn, K.W.; Chen, M.; Green, J.S.; Saad, A.; Antin, J.H.; Savani, B.N.; et al. Early Cytomegalovirus Reactivation Remains Associated with Increased Transplant-Related Mortality in the Current Era: A CIBMTR Analysis. Blood 2016, 127, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, N.; Hirsch, H.H.; Khanna, N.; Gerull, S.; Buser, A.; Bucher, C.; Halter, J.; Heim, D.; Tichelli, A.; Gratwohl, A.; et al. Evidence for a Bidirectional Relationship between Cytomegalovirus Replication and Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2010, 16, 1309–1314. [Google Scholar] [CrossRef] [Green Version]
- Stern, M.; Hirsch, H.; Cusini, A.; van Delden, C.; Manuel, O.; Meylan, P.; Boggian, K.; Mueller, N.J.; Dickenmann, M.; Members of Swiss Transplant Cohort Study. Cytomegalovirus Serology and Replication Remain Associated with Solid Organ Graft Rejection and Graft Loss in the Era of Prophylactic Treatment. Transplantation 2014, 98, 1013–1018. [Google Scholar] [CrossRef] [Green Version]
- Ljungman, P.; de la Camara, R.; Robin, C.; Crocchiolo, R.; Einsele, H.; Hill, J.A.; Hubacek, P.; Navarro, D.; Cordonnier, C.; Ward, K.N. Guidelines for the Management of Cytomegalovirus Infection in Patients with Haematological Malignancies and after Stem Cell Transplantation from the 2017 European Conference on Infections in Leukaemia (ECIL 7). Lancet Infect. Dis. 2019, 19, e260–e272. [Google Scholar] [CrossRef]
- Ljungman, P.; Boeckh, M.; Hirsch, H.H.; Josephson, F.; Lundgren, J.; Nichols, G.; Pikis, A.; Razonable, R.R.; Miller, V.; Griffiths, P.D.; et al. Definitions of Cytomegalovirus Infection and Disease in Transplant Patients for Use in Clinical Trials. Clin. Infect. Dis. 2017, 64, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Fryer, J.F.; Heath, A.B.; Minor, P.D. Collaborative Study Group A Collaborative Study to Establish the 1st WHO International Standard for Human Cytomegalovirus for Nucleic Acid Amplification Technology. Biologicals 2016, 44, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Boom, R.; Sol, C.J.A.; Schuurman, T.; van Breda, A.; Weel, J.F.L.; Beld, M.; ten Berge, I.J.M.; Wertheim-van Dillen, P.M.E.; de Jong, M.D. Human Cytomegalovirus DNA in Plasma and Serum Specimens of Renal Transplant Recipients Is Highly Fragmented. J. Clin. Microbiol. 2002, 40, 4105–4113. [Google Scholar] [CrossRef]
- Tong, Y.; Pang, X.L.; Mabilangan, C.; Preiksaitis, J.K. Determination of the Biological Form of Human Cytomegalovirus DNA in the Plasma of Solid-Organ Transplant Recipients. J. Infect. Dis. 2017, 215, 1094–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiopris, G.; Veronese, P.; Cusenza, F.; Procaccianti, M.; Perrone, S.; Daccò, V.; Colombo, C.; Esposito, S. Congenital Cytomegalovirus Infection: Update on Diagnosis and Treatment. Microorganisms 2020, 8, 1516. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Lee, S.; Lloyd, M.; Irish, A.; Price, P. The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients. IJMS 2019, 20, 5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisboa, L.F.; Åsberg, A.; Kumar, D.; Pang, X.; Hartmann, A.; Preiksaitis, J.K.; Pescovitz, M.D.; Rollag, H.; Jardine, A.G.; Humar, A. The Clinical Utility of Whole Blood Versus Plasma Cytomegalovirus Viral Load Assays for Monitoring Therapeutic Response. Transplantation 2011, 91, 231–236. [Google Scholar] [CrossRef]
- Barrett-Muir, W.; Breuer, J.; Millar, C.; Thomas, J.; Jeffries, D.; Yaqoob, M.; Aitken, C. CMV Viral Load Measurements in Whole Blood and Plasma—Which Is Best Following Renal Transplantation? Transplantation 2000, 70, 116–119. [Google Scholar]
- Garrigue, I.; Boucher, S.; Couzi, L.; Caumont, A.; Dromer, C.; Neau-Cransac, M.; Tabrizi, R.; Schrive, M.-H.; Fleury, H.; Lafon, M.-E. Whole Blood Real-Time Quantitative PCR for Cytomegalovirus Infection Follow-up in Transplant Recipients. J. Clin. Virol. 2006, 36, 72–75. [Google Scholar] [CrossRef]
- Razonable, R.R.; Brown, R.A.; Wilson, J.; Groettum, C.; Kremers, W.; Espy, M.; Smith, T.F.; Paya, C.V. The Clinical Use of Various Blood Compartments for Cytomegalovirus (CMV) DNA Quantitation in Transplant Recipients with CMV Disease. Transplantation 2002, 73, 968–973. [Google Scholar] [CrossRef]
- Tang, W.; Elmore, S.H.; Fan, H.; Thorne, L.B.; Gulley, M.L. Cytomegalovirus DNA Measurement in Blood and Plasma Using Roche LightCycler CMV Quantification Reagents. Diagn. Mol. Pathol. 2008, 17, 166–173. [Google Scholar] [CrossRef]
- Costa, C.; Sidoti, F.; Mantovani, S.; Gregori, G.; Proietti, A.; Ghisetti, V.; Cavallo, R. Comparison of Two Molecular Assays for Detection of Cytomegalovirus DNA in Whole Blood and Plasma Samples from Transplant Recipients. New Microbiol. 2016, 39, 186–191. [Google Scholar]
- Koidl, C.; Bozic, M.; Marth, E.; Kessler, H.H. Detection of CMV DNA: Is EDTA Whole Blood Superior to EDTA Plasma? J. Virol. Methods 2008, 154, 210–212. [Google Scholar] [CrossRef]
- Percivalle, E.; Revello, M.G.; Vago, L.; Morini, F.; Gerna, G. Circulating Endothelial Giant Cells Permissive for Human Cytomegalovirus (HCMV) Are Detected in Disseminated HCMV Infections with Organ Involvement. J. Clin. Investig. 1993, 92, 663–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinuesa, V.; Giménez, E.; Solano, C.; Albert, E.; Torres, I.; Pérez, A.; Hernández-Boluda, J.C.; Piñana, J.L.; Navarro, D. Factors Influencing Cytomegalovirus DNA Load Measurements in Whole Blood and Plasma Specimens from Allogeneic Hematopoietic Stem Cell Transplant Recipients. Diagn. Microbiol. Infect. Dis. 2019, 94, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Babady, N.E.; Cheng, C.; Cumberbatch, E.; Stiles, J.; Papanicolaou, G.; Tang, Y.-W. Monitoring of Cytomegalovirus Viral Loads by Two Molecular Assays in Whole-Blood and Plasma Samples from Hematopoietic Stem Cell Transplant Recipients. J. Clin. Microbiol. 2015, 53, 1252–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzarotto, T.; Chiereghin, A.; Piralla, A.; Piccirilli, G.; Girello, A.; Campanini, G.; Gabrielli, L.; Costa, C.; Prete, A.; Bonifazi, F.; et al. Cytomegalovirus and Epstein-Barr Virus DNA Kinetics in Whole Blood and Plasma of Allogeneic Hematopoietic Stem Cell Transplantation Recipients. Biol. Blood Marrow Transplant. 2018, 24, 1699–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preiksaitis, J.K.; Hayden, R.T.; Tong, Y.; Pang, X.L.; Fryer, J.F.; Heath, A.B.; Cook, L.; Petrich, A.K.; Yu, B.; Caliendo, A.M. Are We There Yet? Impact of the First International Standard for Cytomegalovirus DNA on the Harmonization of Results Reported on Plasma Samples. Clin. Infect. Dis. 2016, 63, 583–589. [Google Scholar] [CrossRef]
- Garrigue, I.; Doussau, A.; Asselineau, J.; Bricout, H.; Couzi, L.; Rio, C.; Merville, P.; Fleury, H.; Lafon, M.-E.; Thiébaut, R. Prediction of Cytomegalovirus (CMV) Plasma Load from Evaluation of CMV Whole-Blood Load in Samples from Renal Transplant Recipients. J. Clin. Microbiol. 2008, 46, 493–498. [Google Scholar] [CrossRef] [Green Version]
- Kraft, C.S.; Armstrong, W.S.; Caliendo, A.M. Interpreting Quantitative Cytomegalovirus DNA Testing: Understanding the Laboratory Perspective. Clin. Infect. Dis. 2012, 54, 1793–1797. [Google Scholar] [CrossRef] [Green Version]
- Razonable, R.R.; Hayden, R.T. Clinical Utility of Viral Load in Management of Cytomegalovirus Infection after Solid Organ Transplantation. Clin. Microbiol. Rev. 2013, 26, 703–727. [Google Scholar] [CrossRef]



| Age Group | Sex | n | Initial Hospitalisation Reason |
|---|---|---|---|
| Children | Female | 11 | Oncology and/or hematology |
| Male | 12 | ||
| Adults | Female | 15 | Solid organ transplantation |
| Male | 15 |
| Whole Blood | Plasma | ||
|---|---|---|---|
| Positive CMV DNA | Positive CMV DNA | Negative CMV DNA | |
| n | 156 | 97 | 59 |
| % | 100 | 62.2 | 37.8 |
| Patient No. | CMV DNA [IU/mL] | |
|---|---|---|
| in Whole Blood | in Plasma | |
| 1. | 2870 | 4155 |
| 2. | 1530 | 5000 |
| 3. | 880 | 2520 |
| 4. | 475 | 3253 |
| 5. | 3180 | 5850 |
| 3100 | 3240 | |
| 5850 | 8500 | |
| 1970 | 3040 | |
| 6. | 1360 | 4325 |
| 7. | 9000 | 18,450 |
| 8. | 123 | 1055 |
| 9. | 172,670 | 188,000 |
| 1950 | 2365 | |
| 10. | 1290 | 10,900 |
| 1320 | 6550 | |
| 4613 | 8700 | |
| 11. | 1,013,340 | 1,585,000 |
| 11,070 | 17,450 | |
| 12. | 27,270 | 27,300 |
| 13. | 45,270 | 88,500 |
| Statistical Analysis | Statistically Significance | |
|---|---|---|
| Correlation of negative plasma result vs. | ||
| χ² = 0.28, p = 0.5945 | No |
| χ² = 0.10, p = 0.7550 | No |
| Correlation of the number of CMV load in the whole blood vs. | Yes | |
| Z = 8.2276, p < 0.001 | |
| Correlation of CMV load in samples positive in both specimens, whole blood and plasma | Spearman’s r = 0.67, p < 0.001 | Yes |
| Correlation of higher value of CMV load in plasma vs. | ||
| χ² = 0.62, p = 0.4309 | No |
| χ² = 0.51, p = 0.4768 | No |
| Median DNAemia values for: | ||
| 4260 IU/mL | |
| 1105 IU/mL | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rzepka, M.; Depka, D.; Gospodarek-Komkowska, E.; Bogiel, T. Whole Blood versus Plasma Samples—How Does the Type of Specimen Collected for Testing Affect the Monitoring of Cytomegalovirus Viremia? Pathogens 2022, 11, 1384. https://doi.org/10.3390/pathogens11111384
Rzepka M, Depka D, Gospodarek-Komkowska E, Bogiel T. Whole Blood versus Plasma Samples—How Does the Type of Specimen Collected for Testing Affect the Monitoring of Cytomegalovirus Viremia? Pathogens. 2022; 11(11):1384. https://doi.org/10.3390/pathogens11111384
Chicago/Turabian StyleRzepka, Mateusz, Dagmara Depka, Eugenia Gospodarek-Komkowska, and Tomasz Bogiel. 2022. "Whole Blood versus Plasma Samples—How Does the Type of Specimen Collected for Testing Affect the Monitoring of Cytomegalovirus Viremia?" Pathogens 11, no. 11: 1384. https://doi.org/10.3390/pathogens11111384