FoMC69 Gene in Fusarium oxysporum f. sp. radicis-lycopersici Is Essential for Pathogenicity by Involving Normal Function of Chlamydospores
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Culture Conditions
2.2. Inoculation Test
2.3. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction
2.4. Fungal Quantification
2.5. Preparation of Gene Deletion and Complementation Constructs
2.6. Fungal Transformation
2.7. Visualizing Fungal Colonization in Planta by WGA-FITC Labeling
2.8. Quantification of Chlamydospores
2.9. Statistical Analysis
3. Results
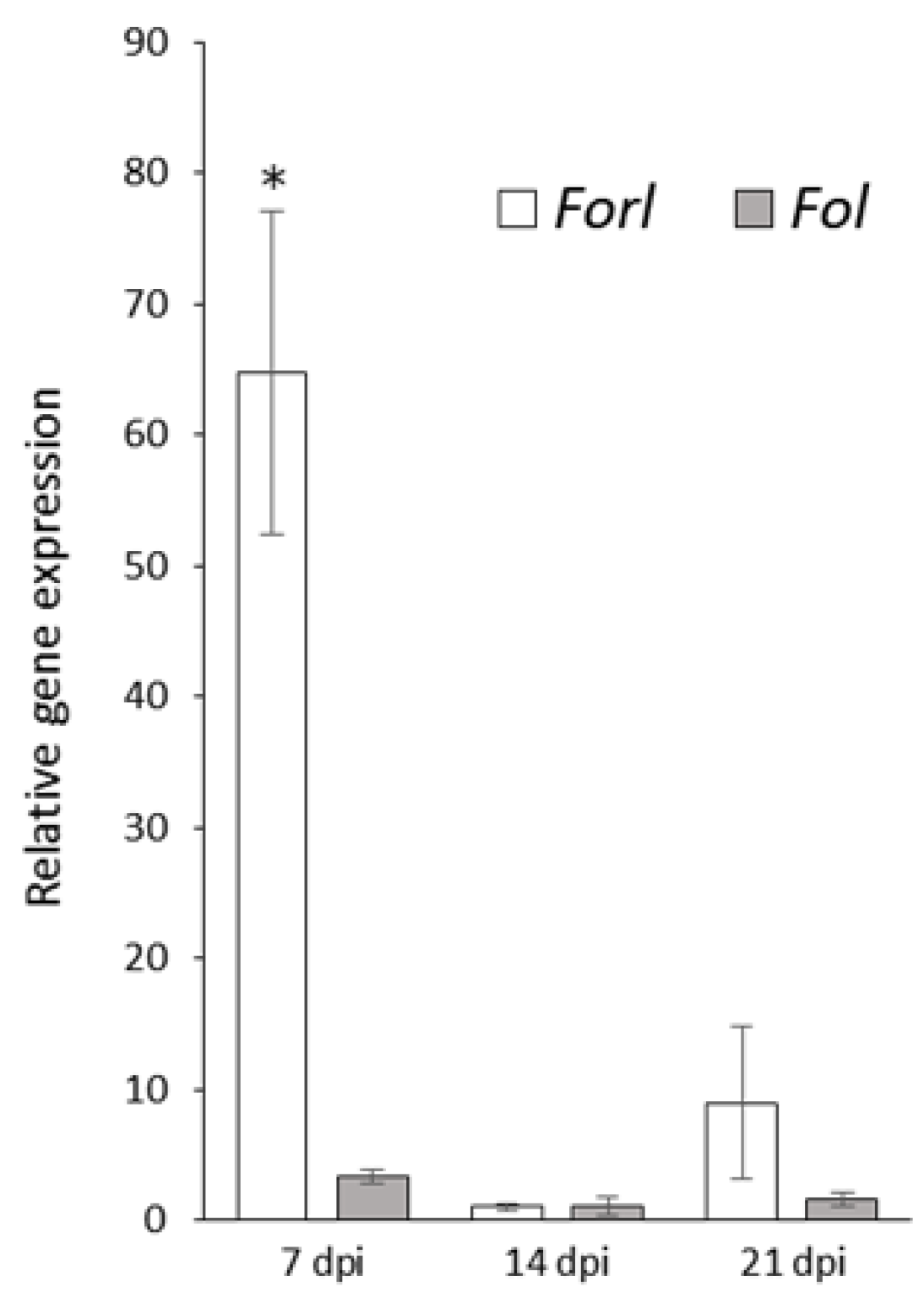
3.1. Characterization of FoMC69 in Forl
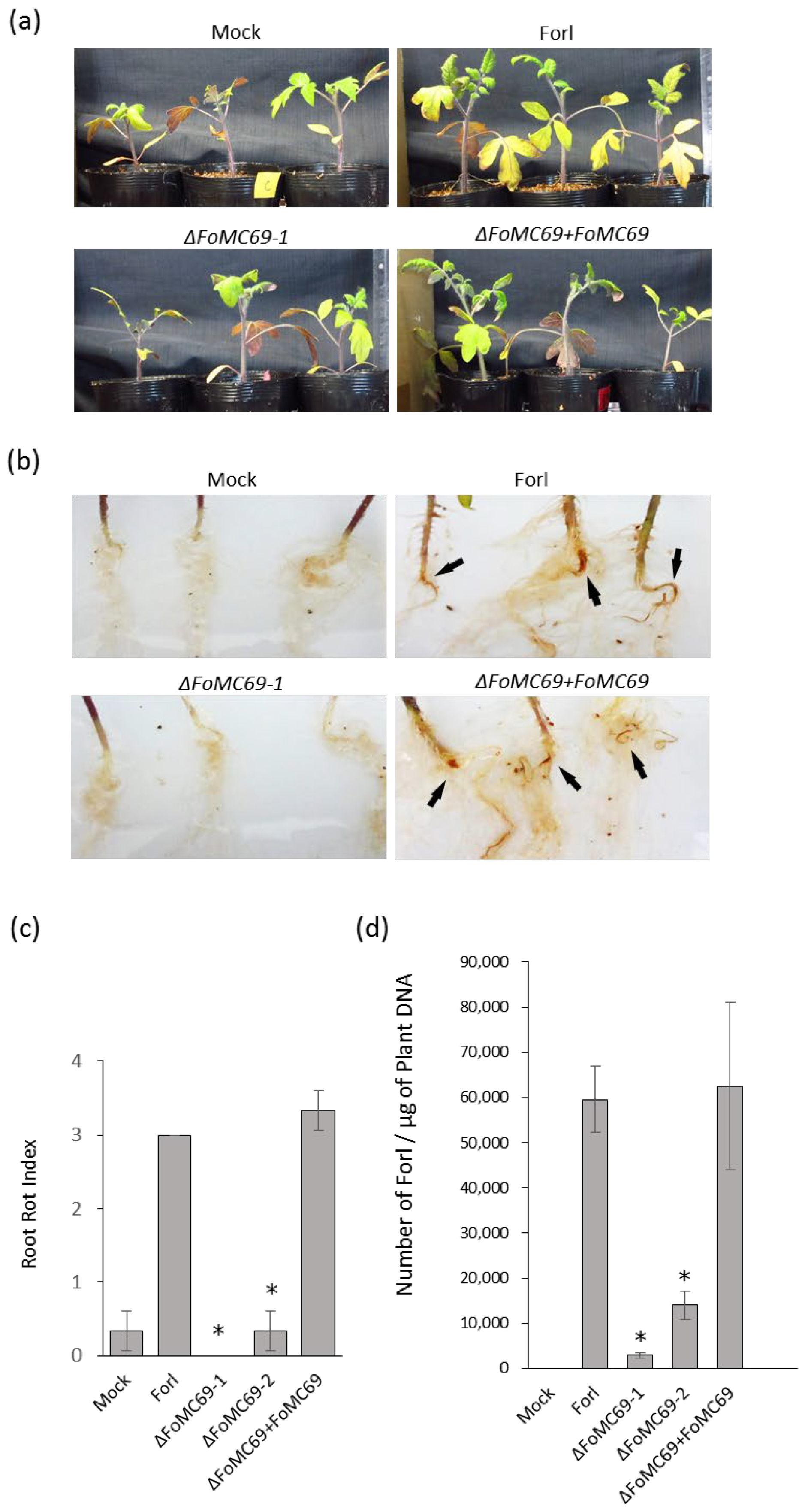
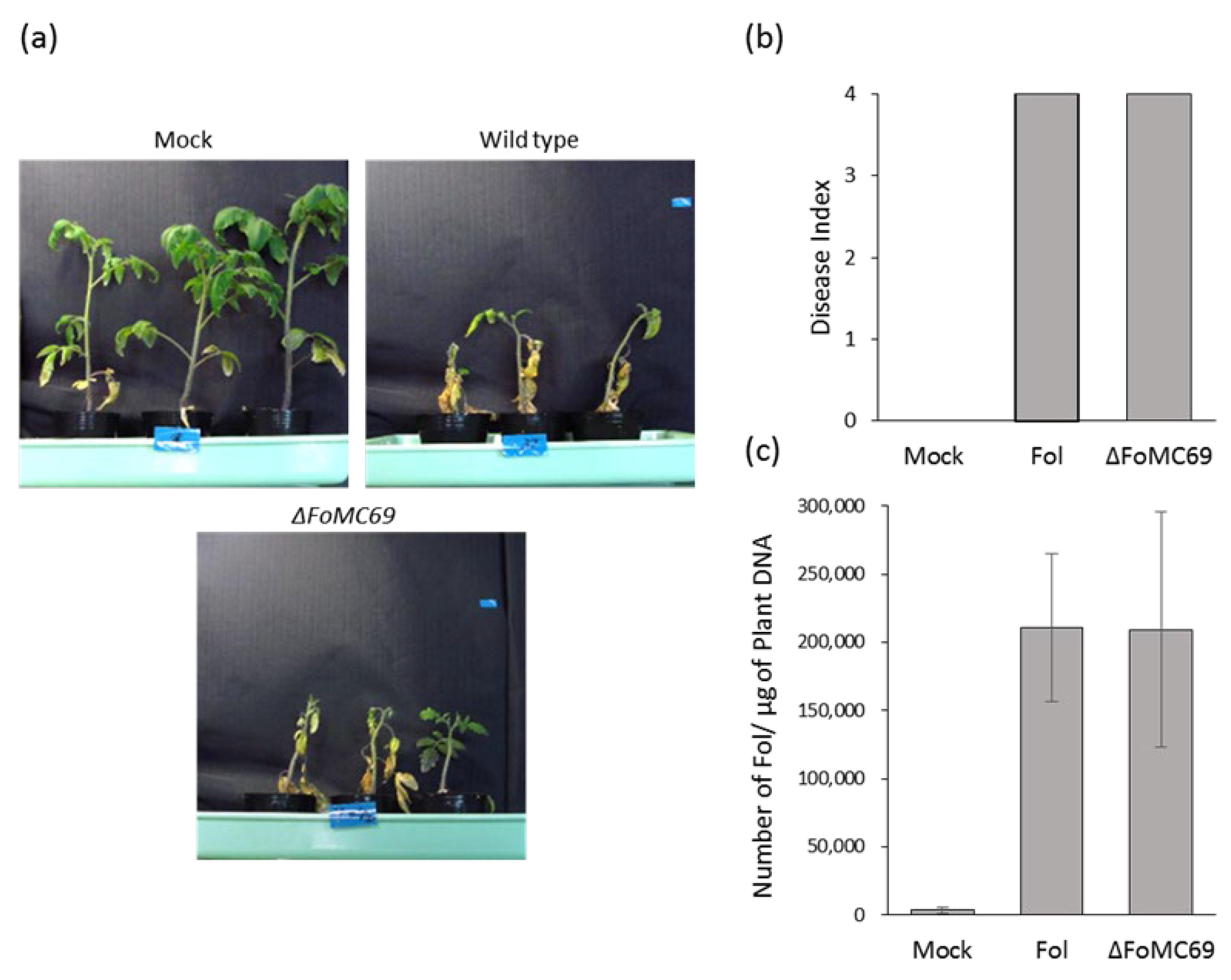
3.2. FoMC69 Involved in Virulence of Forl but Not of Forl
3.3. Morphological Observation and Quantification of Chlamydospore in Planta
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leslie, J.F.; Summerell, B.A. Fusarium laboratory workshops—A recent history. Mycotoxin Res. 2006, 22, 73. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311. [Google Scholar] [CrossRef] [PubMed]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Smolinska, U. Survival of Sclerotium cepivorum sclerotia and Fusarium oxysporum chlamydospores in soil amended with cruciferous residues. J. Phytopathol. 2000, 148, 343–349. [Google Scholar] [CrossRef]
- Smith, S. An overview of ecological and habitat aspects in the genus Fusarium with special emphasis on the soil-borne pathogenic forms. Plant Pathol. Bull. 2007, 16, 97–120. [Google Scholar]
- Kommedahl, T. Relation of exudates of pea roots to germination of spores in races of Fusarium oxysporum f. pisi. Phytopathology 1966, 56, 721–722. [Google Scholar]
- FAO Statistics Division. Available online: http://faostat.fao.org/ (accessed on 8 August 2022).
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
- De Sain, M.; Rep, M. The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 2015, 16, 23970–23993. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.M.; Houterman, P.M.; Schreiver, I.; Ma, L.; Amyotte, S.; Chellappan, B.; Boeren, S.; Takken, F.L.W.; Rep, M. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013, 14, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houterman, P.M.; Speijer, D.; Dekker, H.L.; de Koster, C.G.; Cornelissen, B.J.; Rep, M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007, 8, 215–221. [Google Scholar] [CrossRef]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporumf. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbial. Lett. 2009, 300, 201–215. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef]
- Hemetsberger, C.; Mueller, A.N.; Matei, A.; Herrberger, C.; Hensel, G.; Kumlehn, J.; Mishra, B.; Sharma, R.; Thines, M.; Hückelhoven, R.; et al. The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol. 2015, 206, 1116–1126. [Google Scholar] [CrossRef]
- Saitoh, H.; Fujisawa, S.; Mitsuoka, C.; Ito, A.; Hirabuchi, A.; Ikeda, K.; Irieda, H.; Yoshino, K.; Yoshida, K.; Matsumura, H.; et al. Large-scale gene disruption in Magnaportheoryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PloSPathog. 2012, 8, e1002711. [Google Scholar]
- Eisermann, I.; Weihmann, F.; Krijger, J.J.; Kröling, C.; Hause, G.; Menzel, M.; Pienkny, S.; Kiesoe, A.; Deising, H.D.; Wirsel, S.G. Two genes in a pathogenicity gene cluster encoding secreted proteins are required for appressorial penetration and infection of the maize anthracnose fungus Colletotrichum graminicola. Environ. Microbial. 2019, 21, 4773–4791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namiki, F.; Shiomi, T.; Kayamura, T.; Tsuge, T. Characterization of the formaespeciales of Fusarium oxysporum causing wilt of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 1994, 60, 2684–2691. [Google Scholar] [CrossRef] [Green Version]
- Komada, H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev. Plant Prot. Res. 1975, 8, 114–125. [Google Scholar]
- van der Does, H.C.; Duyvesteijn, R.G.; Goltstein, P.M.; van Schie, C.C.; Manders, E.M.; Cornelissen, B.J.; Rep, M. Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genet. Biol. 2008, 45, 1257–1264. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fujikawa, I.; Takehara, Y.; Ota, M.; Imada, K.; Sasaki, K.; Kajihara, H.; Sakai, S.; Jogaish, S.; Ito, S.I. Magnesium oxide induces immunity against Fusarium wilt by triggering the jasmonic acid signaling pathway in tomato. J. Biotechnol. 2021, 325, 100–108. [Google Scholar] [CrossRef]
- Kuwayama, H.; Obara, S.; Morio, T.; Katoh, M.; Urushihara, H.; Tanaka, Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002, 30, e2. [Google Scholar] [CrossRef]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B.G. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 5, 9–11. [Google Scholar] [CrossRef] [Green Version]
- Namiki, F.; Matsunaga, M.; Okuda, M.; Inoue, I.; Nishi, K.; Fujita, Y.; Tsuge, T. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 2001, 14, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Nakahara, K.; Tanaka, S.; Shigyo, M.; Ito, S. Genetic and pathogenic variability of Fusarium oxysporum f. sp. cepae isolated from onion and Welsh onion in Japan. Phytopathology 2015, 105, 525–532. [Google Scholar] [CrossRef] [Green Version]
- Kawabe, M.; Mizutani, K.; Yoshida, T.; Teraoka, T.; Yoneyama, K.; Yamaguchi, I.; Arie, T. Cloning of the pathogenicity-related gene FPD1 in Fusarium oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 2004, 70, 16–20. [Google Scholar] [CrossRef]
- Saitoh, K.I.; Togashi, K.; Arie, T.; Teraoka, T. A simple method for a mini-preparation of fungal DNA. J. Gen. Plant Pathol. 2006, 72, 348–350. [Google Scholar] [CrossRef]
- Lagopodi, A.L.; Ram, A.F.; Lamers, G.E.; Punt, P.J.; Van den Hondel, C.A.; Lugtenberg, B.J.; Bloemberg, G.V. Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant–Microbe Interact. 2002, 15, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.H.; Hsu, L.H.; Wang, H.F.; Lai, Y.H.; Chen, Y.L. Calcineurin regulates conidiation, chlamydospore formation and virulence in Fusarium oxysporumf. sp. lycopersici. Front. Microbial. 2020, 11, 539702. [Google Scholar] [CrossRef]
- Costa, A.E.S.; da Cunha, F.S.; da Cunha Honorato, A.; Capucho, A.S.; Dias, R.D.C.S.; Borel, J.C.; Ishikawa, F.H. Resistance to Fusarium Wilt in watermelon accessions inoculated by chlamydospores. Sci. Hortic. 2018, 228, 181–186. [Google Scholar] [CrossRef]
- De Cal, A.; Pascual, S.; Melgarejo, P. Infectivity of chlamydospores vs microconidia of Fusarium oxysporum f. sp. lycopersici on tomato. J. Phytopathol. 1997, 145, 231–233. [Google Scholar] [CrossRef]
- Ohara, T.; Inoue, I.; Namiki, F.; Kunoh, H.; Tsuge, T. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics 2004, 166, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Di, X.; Cao, L.; Hughes, R.K.; Tintor, N.; Banfield, M.J.; Takken, F.L. Structure–function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 2017, 216, 897–914. [Google Scholar] [CrossRef] [Green Version]
- Manzo, D.; Ferriello, F.; Puopolo, G.; Zoina, A.; D’Esposito, D.; Tardella, L.; Ferrarini, A.; Ercolano, M.R. Fusarium oxysporum f. sp. radicis-lycopersiciinduces distinct transcriptome reprogramming in resistant and susceptible isogenic tomato lines. BMC Plant Biol. 2016, 16, 53. [Google Scholar] [CrossRef] [Green Version]
- Sharpee, W.; Oh, Y.; Yi, M.; Franck, W.; Eyre, A.; Okagaki, L.H.; Valent, B.; Dean, R.A. Identification and characterization of suppressors of plant cell death (SPD) effectors from Magnaportheoryzae. Mol. Plant Pathol. 2017, 18, 850–863. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasaki, K.; Ito, Y.; Hamada, Y.; Dowaki, A.; Jogaiah, S.; Ito, S.-i. FoMC69 Gene in Fusarium oxysporum f. sp. radicis-lycopersici Is Essential for Pathogenicity by Involving Normal Function of Chlamydospores. Pathogens 2022, 11, 1433. https://doi.org/10.3390/pathogens11121433
Sasaki K, Ito Y, Hamada Y, Dowaki A, Jogaiah S, Ito S-i. FoMC69 Gene in Fusarium oxysporum f. sp. radicis-lycopersici Is Essential for Pathogenicity by Involving Normal Function of Chlamydospores. Pathogens. 2022; 11(12):1433. https://doi.org/10.3390/pathogens11121433
Chicago/Turabian StyleSasaki, Kazunori, Yumi Ito, Yuki Hamada, Ayano Dowaki, Sudisha Jogaiah, and Shin-ichi Ito. 2022. "FoMC69 Gene in Fusarium oxysporum f. sp. radicis-lycopersici Is Essential for Pathogenicity by Involving Normal Function of Chlamydospores" Pathogens 11, no. 12: 1433. https://doi.org/10.3390/pathogens11121433




