Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs
Abstract
:1. Introduction
| Gene Segment (nucleotides *) | Protein | Functions | References |
|---|---|---|---|
| 1 (2313 nt) | PB2 | Recognizes cap structure | [19] |
| 2 (2302 nt) | PB1 | Elongation of RNA synthesis | [25] |
| PB1-F2 | Inhibition of host immune responses | [26,27] | |
| PB1-N40 | Supports the RdRp activity | [2] | |
| 3 (2206 nt) | PA | Endonuclease activity for cap-snatching | [20] |
| PA-X | Host shutoff | [28,29] | |
| PA-N155 | Supports virus replication | [3] | |
| PA-N182 | Supports virus replication | [3] | |
| 4 (1746 nt) | HA | Mediates virus attachment to cells | [5] |
| 5 (1537 nt) | NP | Encapsulates viral genomes | [6,17] |
| 6 (1370 nt) | NA | Facilitates virus release | [30] |
| 7 (996 nt) | M1 | Virion assembly | [8] |
| M2 | Ion channel; virus budding | [9,23] | |
| M42 | Supports virus replication | [10] | |
| 8 (854 nt) | NS1 | Inhibition of host immune responses | [31] |
| NS2 | Nuclear export of vRNPs | [11,22] | |
| NS3 | Host specificity | [12] |
2. Host Gene Shutoff
2.1. Inhibition of Cellular Gene Transcription ((1) in Figure 1)
2.1.1. Degradation of RNA Polymerase II by Viral RdRp
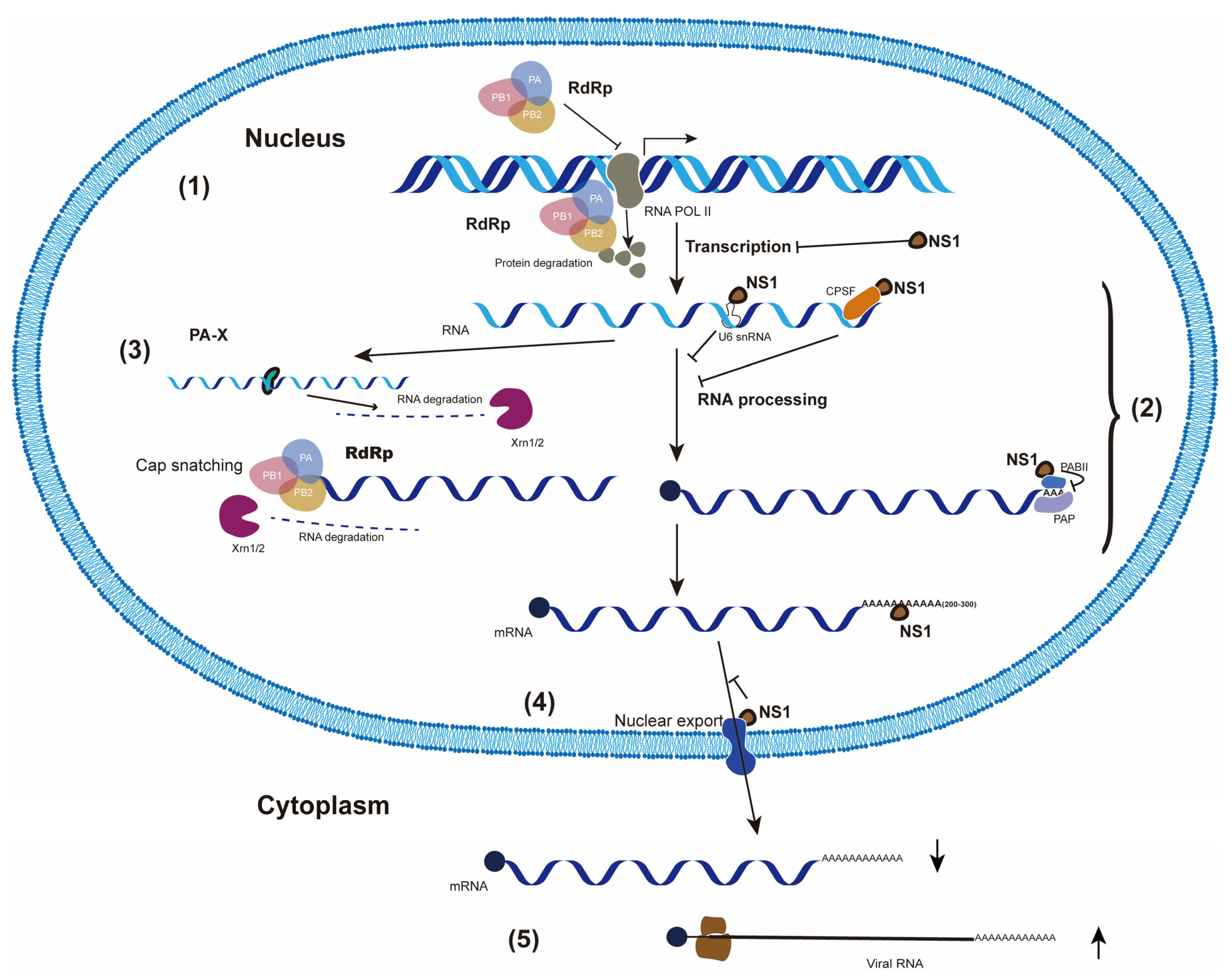
2.1.2. NS1 Inhibits Cellular RNA Transcription Directly
2.2. Suppression of Cellular RNA Processing ((2) in Figure 1)
2.2.1. Cap-Snatching
2.2.2. NS1 Interacts with Cellular pre-mRNA Processing Factors
2.3. Degradation of Cellular mRNAs by PA-X ((3) in Figure 1)
2.4. Blockage of Cellular mRNA in the Nucleus by NS1 ((4) in Figure 1)
2.5. Selective Translation of Viral mRNAs ((5) in Figure 1)
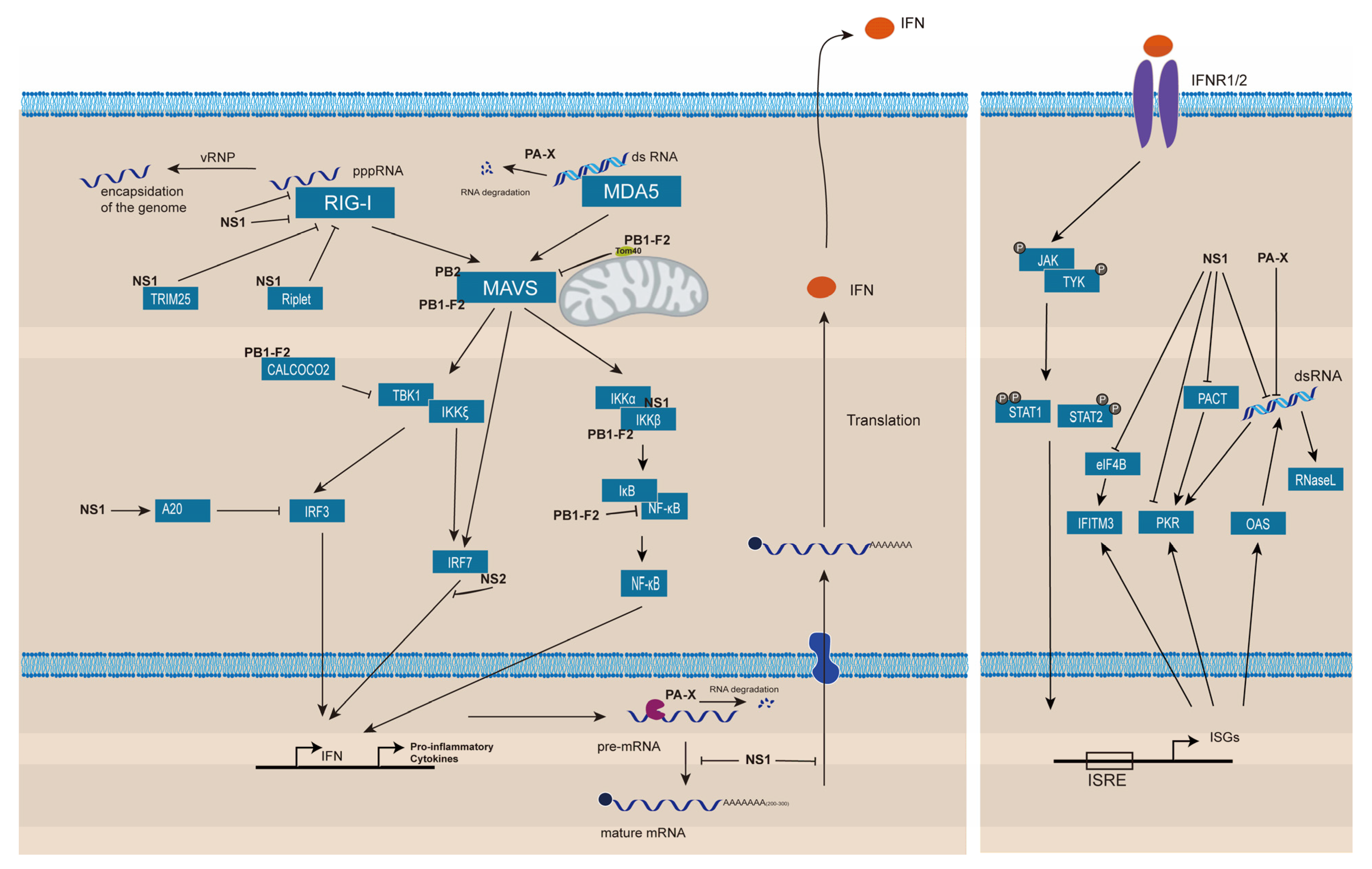
3. Suppression of Host Innate Immune Responses
3.1. Prevention of the Viral RNAs Detection by RLRs
3.2. Inhibition of Proteins Involved in the Signaling Leading to IFN Production
3.3. Shut off of the IFN Gene Expression
3.4. Inhibition of ISG Activities
3.5. Suppression of Inflammasome Formation
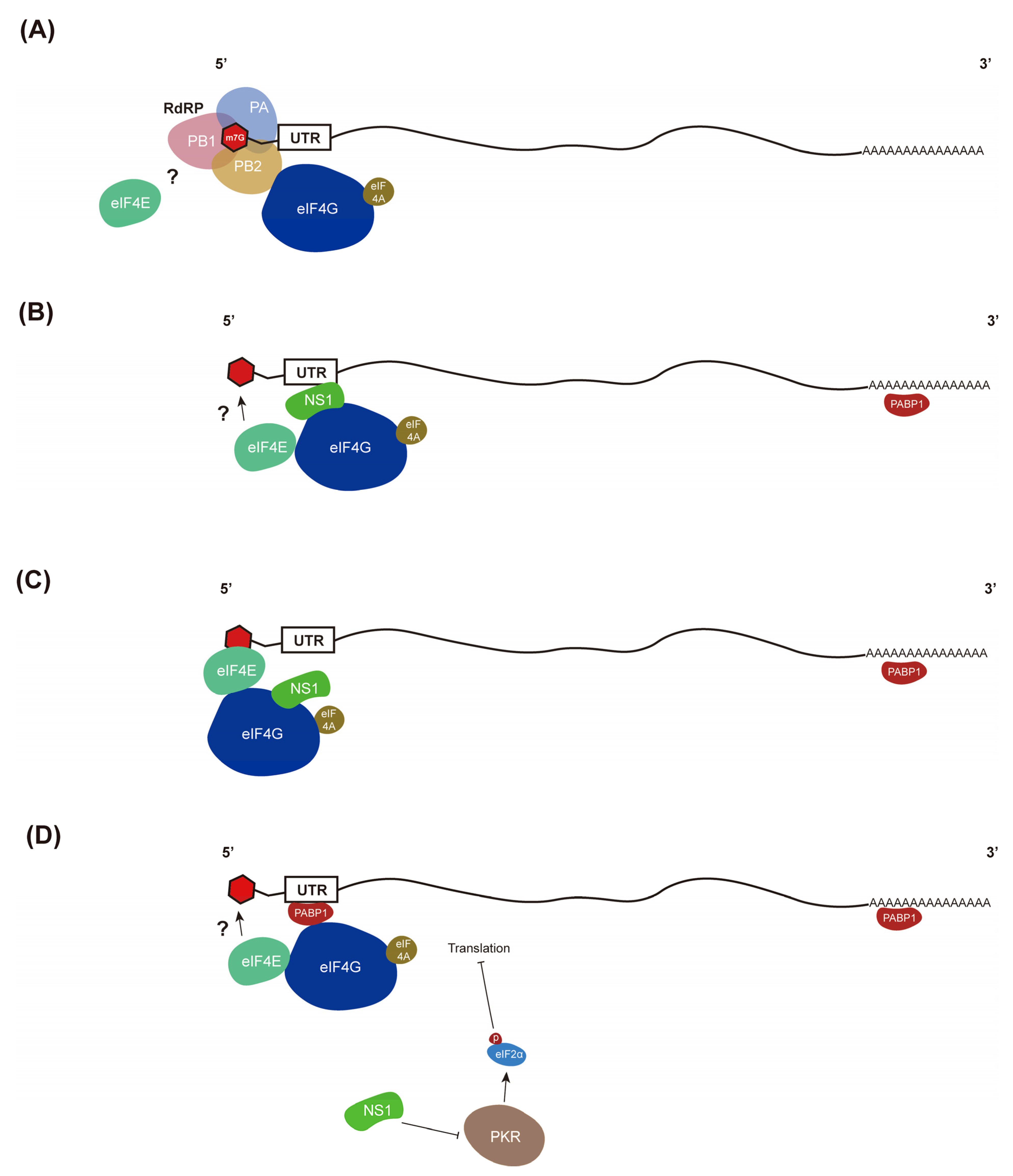
4. Selective Translation of Viral mRNAs
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chauhan, R.P.; Gordon, M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes 2022, 58, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.M.; Foeglein, A.; Sun, J.; Dalton, R.M.; Patel, S.; Howard, W.; Anderson, E.C.; Barclay, W.S.; Digard, P. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 2009, 83, 8021–8031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muramoto, Y.; Noda, T.; Kawakami, E.; Akkina, R.; Kawaoka, Y. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 2013, 87, 2455–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yewdell, J.W.; Ince, W.L. Virology. Frameshifting to PA-X influenza. Science 2012, 337, 164–165. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.Y.; Lin, C.W.; Tsai, T.I.; Lee, C.D.; Chuang, H.Y.; Chen, J.B.; Tsai, M.H.; Chen, B.R.; Lo, P.W.; Liu, C.P.; et al. Influenza A surface glycosylation and vaccine design. Proc. Natl. Acad. Sci. USA 2017, 114, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Bullido, R.; Gomez-Puertas, P.; Albo, C.; Portela, A. Several protein regions contribute to determine the nuclear and cytoplasmic localization of the influenza A virus nucleoprotein. J. Gen. Virol. 2000, 81, 135–142. [Google Scholar] [CrossRef]
- Chen, F.; Liu, T.; Xu, J.; Huang, Y.; Liu, S.; Yang, J. Key amino acid residues of neuraminidase involved in influenza A virus entry. Pathog. Dis. 2019, 77, 6. [Google Scholar] [CrossRef] [Green Version]
- Kordyukova, L.V.; Shtykova, E.V.; Baratova, L.A.; Svergun, D.I.; Batishchev, O.V. Matrix proteins of enveloped viruses: A case study of Influenza A virus M1 protein. J. Biomol. Struct. Dyn. 2019, 37, 671–690. [Google Scholar] [CrossRef]
- Alvarado-Facundo, E.; Gao, Y.; Ribas-Aparicio, R.M.; Jimenez-Alberto, A.; Weiss, C.D.; Wang, W. Influenza virus M2 protein ion channel activity helps to maintain pandemic 2009 H1N1 virus hemagglutinin fusion competence during transport to the cell surface. J. Virol. 2015, 89, 1975–1985. [Google Scholar] [CrossRef] [Green Version]
- Wise, H.M.; Hutchinson, E.C.; Jagger, B.W.; Stuart, A.D.; Kang, Z.H.; Robb, N.; Schwartzman, L.M.; Kash, J.C.; Fodor, E.; Firth, A.E.; et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012, 8, e1002998. [Google Scholar] [CrossRef]
- Paterson, D.; Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 2012, 8, e1003019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selman, M.; Dankar, S.K.; Forbes, N.E.; Jia, J.J.; Brown, E.G. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 2012, 1, e42. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Wang, L.; Li, S. Roles of the Non-Structural Proteins of Influenza A Virus. Pathogens 2020, 9, 10. [Google Scholar] [CrossRef]
- Chatziprodromidou, I.P.; Arvanitidou, M.; Guitian, J.; Apostolou, T.; Vantarakis, G.; Vantarakis, A. Global avian influenza outbreaks 2010–2016: A systematic review of their distribution, avian species and virus subtype. Syst. Rev. 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.M.; Whittaker, G.R. Fusion of Enveloped Viruses in Endosomes. Traffic 2016, 17, 593–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calder, L.J.; Rosenthal, P.B. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat. Struct. Mol. Biol. 2016, 23, 853–858. [Google Scholar] [CrossRef]
- Coloma, R.; Arranz, R.; de la Rosa-Trevin, J.M.; Sorzano, C.O.S.; Munier, S.; Carlero, D.; Naffakh, N.; Ortin, J.; Martin-Benito, J. Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism. Nat. Microbiol. 2020, 5, 727–734. [Google Scholar] [CrossRef]
- Newcomb, L.L.; Kuo, R.L.; Ye, Q.; Jiang, Y.; Tao, Y.J.; Krug, R.M. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 2009, 83, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Guilligay, D.; Tarendeau, F.; Resa-Infante, P.; Coloma, R.; Crepin, T.; Sehr, P.; Lewis, J.; Ruigrok, R.W.; Ortin, J.; Hart, D.J.; et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 2008, 15, 500–506. [Google Scholar] [CrossRef]
- Dias, A.; Bouvier, D.; Crepin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. [Google Scholar] [CrossRef]
- Poon, L.L.; Pritlove, D.C.; Fodor, E.; Brownlee, G.G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 1999, 73, 3473–3476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunotte, L.; Flies, J.; Bolte, H.; Reuther, P.; Vreede, F.; Schwemmle, M. The nuclear export protein of H5N1 influenza A viruses recruits Matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J. Biol. Chem. 2014, 289, 20067–20077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [Green Version]
- Samji, T. Influenza A: Understanding the viral life cycle. Yale J. Biol. Med. 2009, 82, 153–159. [Google Scholar] [PubMed]
- Lin, R.W.; Chen, G.W.; Sung, H.H.; Lin, R.J.; Yen, L.C.; Tseng, Y.L.; Chang, Y.K.; Lien, S.P.; Shih, S.R.; Liao, C.L. Naturally occurring mutations in PB1 affect influenza A virus replication fidelity, virulence, and adaptability. J. Biomed. Sci. 2019, 26, 55. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, A.K.; Pasricha, G. An insight into the PB1F2 protein and its multifunctional role in enhancing the pathogenicity of the influenza A viruses. Virology 2013, 440, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Yoshizumi, T.; Ichinohe, T.; Sasaki, O.; Otera, H.; Kawabata, S.; Mihara, K.; Koshiba, T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014, 5, 4713. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Ma, C.; Liu, X. PA-X: A key regulator of influenza A virus pathogenicity and host immune responses. Med. Microbiol. Immunol. 2018, 207, 255–269. [Google Scholar] [CrossRef]
- Gaucherand, L.; Porter, B.K.; Levene, R.E.; Price, E.L.; Schmaling, S.K.; Rycroft, C.H.; Kevorkian, Y.; McCormick, C.; Khaperskyy, D.A.; Gaglia, M.M. The Influenza A Virus Endoribonuclease PA-X Usurps Host mRNA Processing Machinery to Limit Host Gene Expression. Cell Rep. 2019, 27, 776–792.e7. [Google Scholar] [CrossRef] [Green Version]
- Cohen, M.; Zhang, X.Q.; Senaati, H.P.; Chen, H.W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013, 10, 321. [Google Scholar] [CrossRef]
- Han, C.W.; Jeong, M.S.; Jang, S.B. Structure and Function of the Influenza A Virus Non-Structural Protein 1. J. Microbiol. Biotechnol. 2019, 29, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Gaglia, M.M.; Covarrubias, S.; Wong, W.; Glaunsinger, B.A. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012, 86, 9527–9530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, T.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. Host shutoff activity of VHS and SOX-like proteins: Role in viral survival and immune evasion. Virol. J. 2020, 17, 68. [Google Scholar] [CrossRef] [PubMed]
- Inglis, S.C. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza and herpes simplex virus. Mol. Cell Biol. 1982, 2, 1644–1648. [Google Scholar] [CrossRef] [PubMed]
- Levene, R.E.; Gaglia, M.M. Host Shutoff in Influenza A Virus: Many Means to an End. Viruses 2018, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; Perez-Gonzalez, A.; Nieto, A. Influenza virus infection causes specific degradation of the largest subunit of cellular RNA polymerase II. J. Virol. 2007, 81, 5315–5324. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, O.G.; Smith, M.; Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 2005, 79, 5812–5818. [Google Scholar] [CrossRef] [Green Version]
- Llompart, C.M.; Nieto, A.; Rodriguez-Frandsen, A. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. J. Virol. 2014, 88, 3455–3463. [Google Scholar] [CrossRef] [Green Version]
- Vreede, F.T.; Chan, A.Y.; Sharps, J.; Fodor, E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 2010, 396, 125–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, A.Y.; Vreede, F.T.; Smith, M.; Engelhardt, O.G.; Fodor, E. Influenza virus inhibits RNA polymerase II elongation. Virology 2006, 351, 210–217. [Google Scholar] [CrossRef]
- Bauer, D.L.V.; Tellier, M.; Martinez-Alonso, M.; Nojima, T.; Proudfoot, N.J.; Murphy, S.; Fodor, E. Influenza Virus Mounts a Two-Pronged Attack on Host RNA Polymerase II Transcription. Cell Rep. 2018, 23, 2119–2129.e3. [Google Scholar] [CrossRef] [PubMed]
- Vreede, F.T.; Fodor, E. The role of the influenza virus RNA polymerase in host shut-off. Virulence 2010, 1, 436–439. [Google Scholar] [CrossRef]
- Rodriguez, A.; Perez-Gonzalez, A.; Hossain, M.J.; Chen, L.M.; Rolling, T.; Perez-Brena, P.; Donis, R.; Kochs, G.; Nieto, A. Attenuated strains of influenza A viruses do not induce degradation of RNA polymerase II. J. Virol. 2009, 83, 11166–11174. [Google Scholar] [CrossRef] [Green Version]
- Marazzi, I.; Ho, J.S.; Kim, J.; Manicassamy, B.; Dewell, S.; Albrecht, R.A.; Seibert, C.W.; Schaefer, U.; Jeffrey, K.L.; Prinjha, R.K.; et al. Suppression of the antiviral response by an influenza histone mimic. Nature 2012, 483, 428–433. [Google Scholar] [CrossRef] [Green Version]
- Nacken, W.; Schreiber, A.; Masemann, D.; Ludwig, S. The Effector Domain of the Influenza A Virus Nonstructural Protein NS1 Triggers Host Shutoff by Mediating Inhibition and Global Deregulation of Host Transcription When Associated with Specific Structures in the Nucleus. mBio 2021, 12, e0219621. [Google Scholar] [CrossRef]
- Gu, W.; Gallagher, G.R.; Dai, W.; Liu, P.; Li, R.; Trombly, M.I.; Gammon, D.B.; Mello, C.C.; Wang, J.P.; Finberg, R.W. Influenza A virus preferentially snatches noncoding RNA caps. RNA 2015, 21, 2067–2075. [Google Scholar] [CrossRef] [Green Version]
- Nemeroff, M.E.; Barabino, S.M.; Li, Y.; Keller, W.; Krug, R.M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol. Cell 1998, 1, 991–1000. [Google Scholar] [CrossRef]
- Kuo, R.L.; Krug, R.M. Influenza a virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 2009, 83, 1611–1616. [Google Scholar] [CrossRef] [Green Version]
- Mazel-Sanchez, B.; Iwaszkiewicz, J.; Bonifacio, J.P.P.; Silva, F.; Niu, C.; Strohmeier, S.; Eletto, D.; Krammer, F.; Tan, G.; Zoete, V.; et al. Influenza A viruses balance ER stress with host protein synthesis shutoff. Proc. Natl. Acad. Sci. USA 2021, 118, 36. [Google Scholar] [CrossRef]
- Kuo, R.L.; Chen, C.J.; Tam, E.H.; Huang, C.G.; Li, L.H.; Li, Z.H.; Su, P.C.; Liu, H.P.; Wu, C.C. Interactome Analysis of NS1 Protein Encoded by Influenza A H7N9 Virus Reveals an Inhibitory Role of NS1 in Host mRNA Maturation. J. Proteome Res. 2018, 17, 1474–1484. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Krug, R.M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3’-end processing machinery. EMBO J. 1999, 18, 2273–2283. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Wambach, M.; Katze, M.G.; Krug, R.M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 1995, 214, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, K.; Iguchi, A.; Gomyou, R.; Ono, Y. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 1999, 254, 213–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, P.L.; Chiou, N.T.; Kuss, S.; Garcia-Sastre, A.; Lynch, K.W.; Fontoura, B.M. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 2013, 9, e1003460. [Google Scholar] [CrossRef] [PubMed]
- Beloso, A.; Martinez, C.; Valcarcel, J.; Santaren, J.F.; Ortin, J. Degradation of cellular mRNA during influenza virus infection: Its possible role in protein synthesis shutoff. J. Gen. Virol. 1992, 73, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Jagger, B.W.; Wise, H.M.; Kash, J.C.; Walters, K.A.; Wills, N.M.; Xiao, Y.L.; Dunfee, R.L.; Schwartzman, L.M.; Ozinsky, A.; Bell, G.L.; et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 2012, 337, 199–204. [Google Scholar] [CrossRef] [Green Version]
- Firth, A.E.; Jagger, B.W.; Wise, H.M.; Nelson, C.C.; Parsawar, K.; Wills, N.M.; Napthine, S.; Taubenberger, J.K.; Digard, P.; Atkins, J.F. Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol. 2012, 2, 120109. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Jagger, B.W.; Wise, H.M.; Digard, P.; Holmes, E.C.; Taubenberger, J.K. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 2012, 86, 12411–12413. [Google Scholar] [CrossRef] [Green Version]
- Datta, K.; Wolkerstorfer, A.; Szolar, O.H.; Cusack, S.; Klumpp, K. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 2013, 41, 8289–8299. [Google Scholar] [CrossRef]
- Desmet, E.A.; Bussey, K.A.; Stone, R.; Takimoto, T. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J. Virol. 2013, 87, 3108–3118. [Google Scholar] [CrossRef]
- Oishi, K.; Yamayoshi, S.; Kawaoka, Y. Mapping of a Region of the PA-X Protein of Influenza A Virus That Is Important for Its Shutoff Activity. J. Virol. 2015, 89, 8661–8665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaperskyy, D.A.; Schmaling, S.; Larkins-Ford, J.; McCormick, C.; Gaglia, M.M. Selective Degradation of Host RNA Polymerase II Transcripts by Influenza A Virus PA-X Host Shutoff Protein. PLoS Pathog. 2016, 12, e1005427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins. Viruses 2018, 10, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levene, R.E.; Shrestha, S.D.; Gaglia, M.M. The influenza A virus host shutoff factor PA-X is rapidly turned over in a strain-specific manner. J. Virol. 2021, 95, e02312-20. [Google Scholar] [CrossRef]
- Oishi, K.; Yamayoshi, S.; Kozuka-Hata, H.; Oyama, M.; Kawaoka, Y. N-Terminal Acetylation by NatB Is Required for the Shutoff Activity of Influenza A Virus PA-X. Cell Rep. 2018, 24, 851–860. [Google Scholar] [CrossRef] [Green Version]
- Chaimayo, C.; Dunagan, M.; Hayashi, T.; Santoso, N.; Takimoto, T. Specificity and functional interplay between influenza virus PA-X and NS1 shutoff activity. PLoS Pathog. 2018, 14, e1007465. [Google Scholar] [CrossRef]
- Kuss, S.K.; Mata, M.A.; Zhang, L.; Fontoura, B.M. Nuclear imprisonment: Viral strategies to arrest host mRNA nuclear export. Viruses 2013, 5, 1824–1849. [Google Scholar] [CrossRef]
- Satterly, N.; Tsai, P.L.; van Deursen, J.; Nussenzveig, D.R.; Wang, Y.; Faria, P.A.; Levay, A.; Levy, D.E.; Fontoura, B.M. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2007, 104, 1853–1858. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Xie, Y.; Munoz-Moreno, R.; Wang, J.; Zhang, L.; Esparza, M.; Garcia-Sastre, A.; Fontoura, B.M.A.; Ren, Y. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat. Microbiol. 2019, 4, 1671–1679. [Google Scholar] [CrossRef]
- Pereira, C.F.; Read, E.K.C.; Wise, H.M.; Amorim, M.J.; Digard, P. Influenza A Virus NS1 Protein Promotes Efficient Nuclear Export of Unspliced Viral M1 mRNA. J. Virol. 2017, 91, 15. [Google Scholar] [CrossRef]
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Rodriguez, L.; DeDiego, M.L.; Topham, D.J.; Martinez-Sobrido, L. Interplay of PA-X and NS1 Proteins in Replication and Pathogenesis of a Temperature-Sensitive 2009 Pandemic H1N1 Influenza A Virus. J. Virol. 2017, 91, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Kell, A.M.; Gale, M., Jr. RIG-I in RNA virus recognition. Virology 2015, 479–480, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Hinay, A.A., Jr.; Kakee, S.; Kageyama, S.; Tsuneki-Tokunaga, A.; Perdana, W.Y.; Akena, Y.; Nishiyama, S.; Kanai, K. Pro-Inflammatory Cytokines and Interferon-Stimulated Gene Responses Induced by Seasonal Influenza A Virus with Varying Growth Capabilities in Human Lung Epithelial Cell Lines. Vaccines 2022, 10, 9. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [Green Version]
- Dunagan, M.M.; Hardy, K.; Takimoto, T. Impact of Influenza A Virus Shutoff Proteins on Host Immune Responses. Vaccines 2021, 9, 6. [Google Scholar] [CrossRef]
- Malik, G.; Zhou, Y. Innate Immune Sensing of Influenza A Virus. Viruses 2020, 12, 7. [Google Scholar] [CrossRef]
- Klemm, C.; Boergeling, Y.; Ludwig, S.; Ehrhardt, C. Immunomodulatory Nonstructural Proteins of Influenza A Viruses. Trends Microbiol. 2018, 26, 624–636. [Google Scholar] [CrossRef]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef]
- Zell, R.; Krumbholz, A.; Eitner, A.; Krieg, R.; Halbhuber, K.J.; Wutzler, P. Prevalence of PB1-F2 of influenza A viruses. J. Gen. Virol. 2007, 88, 536–546. [Google Scholar] [CrossRef]
- Weber, M.; Sediri, H.; Felgenhauer, U.; Binzen, I.; Banfer, S.; Jacob, R.; Brunotte, L.; Garcia-Sastre, A.; Schmid-Burgk, J.L.; Schmidt, T.; et al. Influenza virus adaptation PB2-627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 2015, 17, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Naslund, T.I.; Liljestrom, P.; Weber, F.; Reis e Sousa, C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 2006, 314, 997–1001. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Hayashi, T.; MacDonald, L.A.; Takimoto, T. Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses. J. Virol. 2015, 89, 6442–6452. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, J.; Munoz-Moreno, R.; Kim, M.; Sakthivel, R.; Mo, W.; Shao, D.; Anantharaman, A.; Garcia-Sastre, A.; Conrad, N.K.; et al. Influenza Virus NS1 Protein-RNA Interactome Reveals Intron Targeting. J. Virol. 2018, 92, 24. [Google Scholar] [CrossRef]
- Oshiumi, H.; Matsumoto, M.; Seya, T. Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I. J. Biochem. 2012, 151, 5–11. [Google Scholar] [CrossRef]
- Rajsbaum, R.; Albrecht, R.A.; Wang, M.K.; Maharaj, N.P.; Versteeg, G.A.; Nistal-Villan, E.; Garcia-Sastre, A.; Gack, M.U. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012, 8, e1003059. [Google Scholar] [CrossRef]
- Koshiba, T.; Yasukawa, K.; Yanagi, Y.; Kawabata, S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal 2011, 4, ra7. [Google Scholar] [CrossRef]
- Varga, Z.T.; Grant, A.; Manicassamy, B.; Palese, P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J. Virol. 2012, 86, 8359–8366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graef, K.M.; Vreede, F.T.; Lau, Y.F.; McCall, A.W.; Carr, S.M.; Subbarao, K.; Fodor, E. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 2010, 84, 8433–8445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Song, L.; Li, J.; Zhang, Z.; Peng, H.; Jiang, W.; Wang, Q.; Kang, T.; Chen, S.; Huang, W. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol. 2012, 14, 1849–1866. [Google Scholar] [CrossRef]
- Reis, A.L.; McCauley, J.W. The influenza virus protein PB1-F2 interacts with IKKbeta and modulates NF-kappaB signalling. PLoS ONE 2013, 8, e63852. [Google Scholar] [CrossRef] [Green Version]
- Leymarie, O.; Meyer, L.; Tafforeau, L.; Lotteau, V.; Costa, B.D.; Delmas, B.; Chevalier, C.; Le Goffic, R. Influenza virus protein PB1-F2 interacts with CALCOCO2 (NDP52) to modulate innate immune response. J. Gen. Virol. 2017, 98, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Yamamoto, M.; Miyagishi, M.; Taira, K.; Nakanishi, M.; Fujita, T.; Akira, S.; Yamamoto, N.; Yamaoka, S. A20 is a negative regulator of IFN regulatory factor 3 signaling. J. Immunol. 2005, 174, 1507–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, W.; Sun, X.; Shi, N.; Zhang, M.; Guan, Z.; Duan, M. Influenza a virus NS1 protein induced A20 contributes to viral replication by suppressing interferon-induced antiviral response. Biochem. Biophys. Res. Commun. 2017, 482, 1107–1113. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, M.; Huang, J.; Zeng, Q.; Zhu, Q.; Xu, S.; Chen, H. H1N1 Influenza A Virus Protein NS2 Inhibits Innate Immune Response by Targeting IRF7. Viruses 2022, 14, 11. [Google Scholar] [CrossRef]
- Rigby, R.E.; Wise, H.M.; Smith, N.; Digard, P.; Rehwinkel, J. PA-X antagonises MAVS-dependent accumulation of early type I interferon messenger RNAs during influenza A virus infection. Sci. Rep. 2019, 9, 7216. [Google Scholar] [CrossRef] [Green Version]
- Min, J.Y.; Krug, R.M. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2’-5’ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 7100–7105. [Google Scholar] [CrossRef]
- Patel, R.C.; Sen, G.C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998, 17, 4379–4390. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.L.; Katze, M.G. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 1998, 18, 757–766. [Google Scholar] [CrossRef]
- Wang, S.; Chi, X.; Wei, H.; Chen, Y.; Chen, Z.; Huang, S.; Chen, J.L. Influenza A virus-induced degradation of eukaryotic translation initiation factor 4B contributes to viral replication by suppressing IFITM3 protein expression. J. Virol. 2014, 88, 8375–8385. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef] [Green Version]
- Park, H.S.; Liu, G.; Thulasi Raman, S.N.; Landreth, S.L.; Liu, Q.; Zhou, Y. NS1 Protein of 2009 Pandemic Influenza A Virus Inhibits Porcine NLRP3 Inflammasome-Mediated Interleukin-1 Beta Production by Suppressing ASC Ubiquitination. J. Virol. 2018, 92, 8. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, M.; Chen, I.Y.; Kawaguchi, A.; Koshiba, T.; Nagata, K.; Takeyama, H.; Hasegawa, H.; Ichinohe, T. The RNA- and TRIM25-Binding Domains of Influenza Virus NS1 Protein Are Essential for Suppression of NLRP3 Inflammasome-Mediated Interleukin-1beta Secretion. J. Virol. 2016, 90, 4105–4114. [Google Scholar] [CrossRef] [Green Version]
- Brito Querido, J.; Sokabe, M.; Kraatz, S.; Gordiyenko, Y.; Skehel, J.M.; Fraser, C.S.; Ramakrishnan, V. Structure of a human 48S translational initiation complex. Science 2020, 369, 1220–1227. [Google Scholar] [CrossRef]
- Yanguez, E.; Nieto, A. So similar, yet so different: Selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell. Virus Res. 2011, 156, 1–12. [Google Scholar] [CrossRef]
- Katze, M.G.; Krug, R.M. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol. Cell Biol. 1984, 4, 2198–2206. [Google Scholar]
- Feigenblum, D.; Schneider, R.J. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J. Virol. 1993, 67, 3027–3035. [Google Scholar] [CrossRef] [Green Version]
- Burgui, I.; Yanguez, E.; Sonenberg, N.; Nieto, A. Influenza virus mRNA translation revisited: Is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007, 81, 12427–12438. [Google Scholar] [CrossRef]
- Yanguez, E.; Castello, A.; Welnowska, E.; Carrasco, L.; Goodfellow, I.; Nieto, A. Functional impairment of eIF4A and eIF4G factors correlates with inhibition of influenza virus mRNA translation. Virology 2011, 413, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Yanguez, E.; Rodriguez, P.; Goodfellow, I.; Nieto, A. Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 2012, 422, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Bier, K.; York, A.; Fodor, E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 2011, 92, 1627–1634. [Google Scholar] [CrossRef]
- Garfinkel, M.S.; Katze, M.G. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5’-untranslated region. J. Biol. Chem. 1993, 268, 22223–22226. [Google Scholar] [CrossRef]
- Park, Y.W.; Katze, M.G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 1995, 270, 28433–28439. [Google Scholar] [CrossRef] [Green Version]
- De la Luna, S.; Fortes, P.; Beloso, A.; Ortin, J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 1995, 69, 2427–2433. [Google Scholar] [CrossRef] [Green Version]
- Aragon, T.; de la Luna, S.; Novoa, I.; Carrasco, L.; Ortin, J.; Nieto, A. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 2000, 20, 6259–6268. [Google Scholar] [CrossRef]
- Burgui, I.; Aragon, T.; Ortin, J.; Nieto, A. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 2003, 84, 3263–3274. [Google Scholar] [CrossRef]
- Arias-Mireles, B.H.; de Rozieres, C.M.; Ly, K.; Joseph, S. RNA Modulates the Interaction between Influenza A Virus NS1 and Human PABP1. Biochemistry 2018, 57, 3590–3598. [Google Scholar] [CrossRef]
- Cruz, A.; Joseph, S. Interaction of the Influenza A Virus NS1 Protein with the 5’-m7G-mRNA.eIF4E.eIF4G1 Complex. Biochemistry 2022, 61, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- De Rozieres, C.M.; Pequeno, A.; Shahabi, S.; Lucas, T.M.; Godula, K.; Ghosh, G.; Joseph, S. PABP1 Drives the Selective Translation of Influenza A Virus mRNA. J. Mol. Biol. 2022, 434, 167460. [Google Scholar] [CrossRef] [PubMed]
- De Rozieres, C.M.; Joseph, S. Influenza A Virus NS1 Protein Binds as a Dimer to RNA-Free PABP1 but Not to the PABP1.Poly(A) RNA Complex. Biochemistry 2020, 59, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sastre, A.; Egorov, A.; Matassov, D.; Brandt, S.; Levy, D.E.; Durbin, J.E.; Palese, P.; Muster, T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 1998, 252, 324–330. [Google Scholar] [CrossRef] [Green Version]
- Tran, V.; Ledwith, M.P.; Thamamongood, T.; Higgins, C.A.; Tripathi, S.; Chang, M.W.; Benner, C.; Garcia-Sastre, A.; Schwemmle, M.; Boon, A.C.M.; et al. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat. Microbiol. 2020, 5, 1490–1503. [Google Scholar] [CrossRef]
- Hayashi, T.; Chaimayo, C.; McGuinness, J.; Takimoto, T. Critical Role of the PA-X C-Terminal Domain of Influenza A Virus in Its Subcellular Localization and Shutoff Activity. J. Virol. 2016, 90, 7131–7141. [Google Scholar] [CrossRef] [Green Version]
- Salvatore, M.; Basler, C.F.; Parisien, J.P.; Horvath, C.M.; Bourmakina, S.; Zheng, H.; Muster, T.; Palese, P.; Garcia-Sastre, A. Effects of influenza A virus NS1 protein on protein expression: The NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 2002, 76, 1206–1212. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Hsu, C.F.; Lai, X.Q.; Chan, Y.R.; Li, H.C.; Lo, S.Y. Cellular PSMB4 Protein Suppresses Influenza A Virus Replication through Targeting NS1 Protein. Viruses 2022, 14, 10. [Google Scholar] [CrossRef]
- Nogales, A.; Martinez-Sobrido, L.; Chiem, K.; Topham, D.J.; DeDiego, M.L. Functional Evolution of the 2009 Pandemic H1N1 Influenza Virus NS1 and PA in Humans. J. Virol. 2018, 92, 19. [Google Scholar] [CrossRef] [Green Version]
- Marion, R.M.; Aragon, T.; Beloso, A.; Nieto, A.; Ortin, J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997, 25, 4271–4277. [Google Scholar] [CrossRef] [Green Version]
- Min, J.Y.; Li, S.; Sen, G.C.; Krug, R.M. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 2007, 363, 236–243. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.-C.; Yang, C.-H.; Lo, S.-Y. Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs. Pathogens 2022, 11, 1521. https://doi.org/10.3390/pathogens11121521
Li H-C, Yang C-H, Lo S-Y. Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs. Pathogens. 2022; 11(12):1521. https://doi.org/10.3390/pathogens11121521
Chicago/Turabian StyleLi, Hui-Chun, Chee-Hing Yang, and Shih-Yen Lo. 2022. "Strategies of Influenza A Virus to Ensure the Translation of Viral mRNAs" Pathogens 11, no. 12: 1521. https://doi.org/10.3390/pathogens11121521






