A Systemic Study of Subcellular Localization of Porcine Epidemic Diarrhea Virus Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Plasmids and Antibodies
2.2. Molecular Cloning
2.3. Immunofluorescence Assay
2.4. Confocal Microscopy
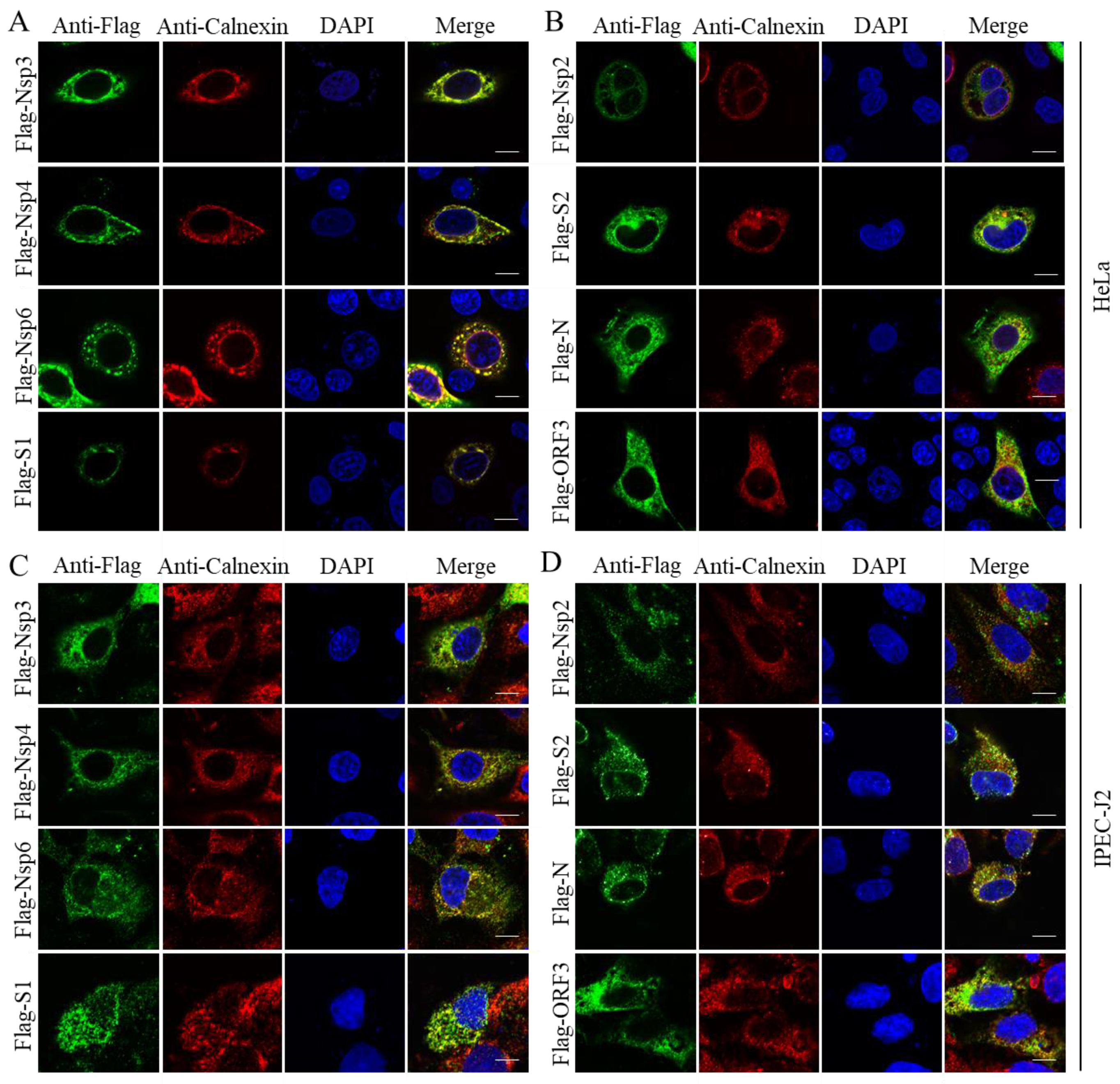
3. Results
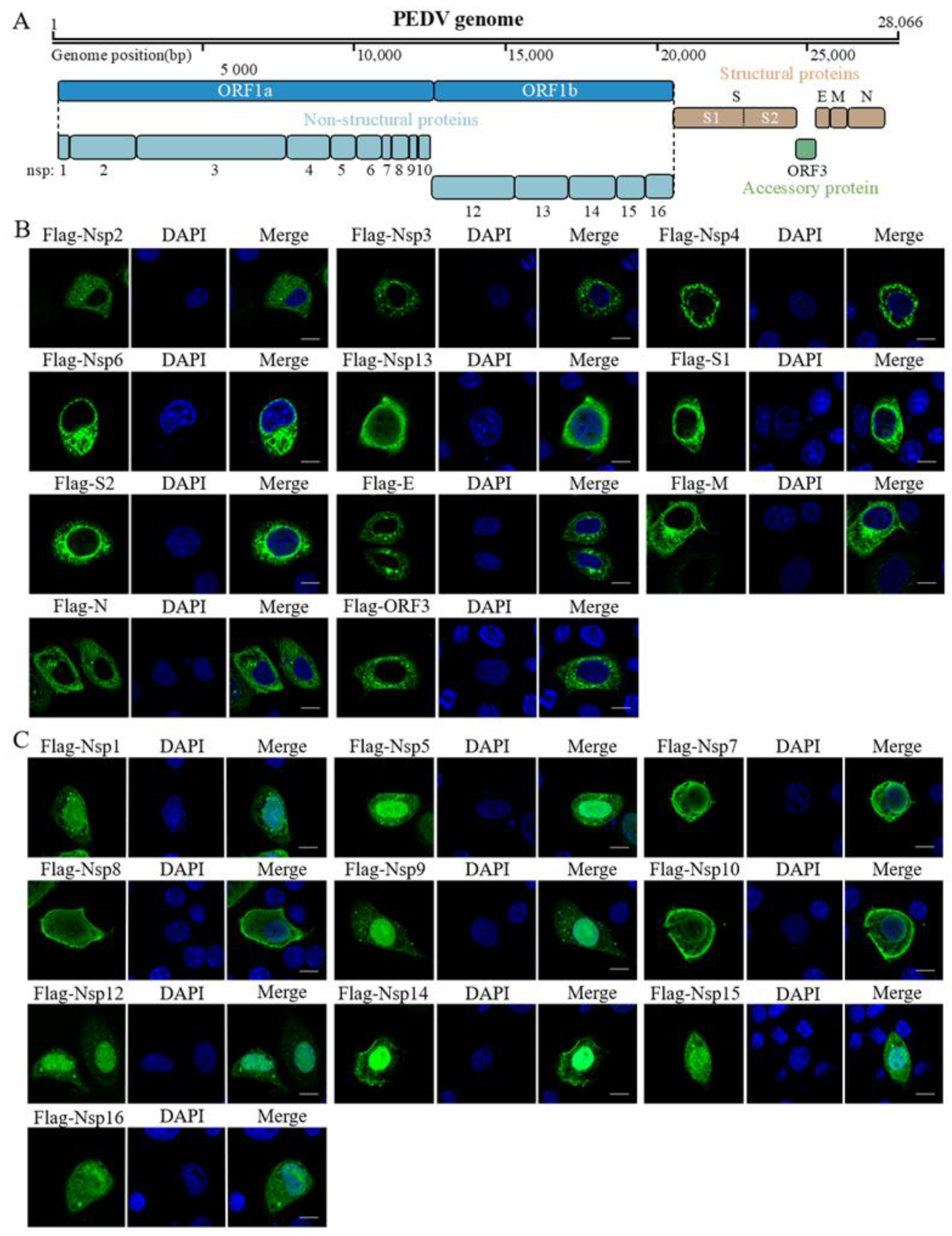
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 204, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New Variants of Porcine Epidemic Diarrhea Virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Cai, R.; Chen, Y.; Liang, P.; Chen, D.; Song, C. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Lee, S.; Lee, C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014, 20, 1223–1226. [Google Scholar] [CrossRef]
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef]
- Duarte, M.; Gelfi, J.; Lambert, P.; Rasschaert, D.; Laude, H. Genome Organization of Porcine Epidemic Diarrhea Virus. In Proceedings of the 5th International Symposium on Coronaviruses, Chantilly, France, 13–18 September 1992; pp. 55–60. [Google Scholar]
- Kamitani, W.; Narayanan, K.; Huang, C.; Lokugamage, K.; Ikegami, T.; Ito, N.; Kubo, H.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 12885–12890. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ke, H.; Blikslager, A.; Fujita, T.; Yoo, D. Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling. J. Virol. 2018, 92, e01677-17. [Google Scholar] [CrossRef]
- Yang, X.; Chen, X.; Bian, G.; Tu, J.; Xing, Y.; Wang, Y.; Chen, Z. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. Gen. Virol. 2014, 95, 614–626. [Google Scholar] [CrossRef]
- Xing, Y.; Chen, J.; Tu, J.; Zhang, B.; Chen, X.; Shi, H.; Baker, S.; Feng, L.; Chen, Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013, 94, 1554–1567. [Google Scholar] [CrossRef]
- Wang, D.; Fang, L.; Shi, Y.; Zhang, H.; Gao, L.; Peng, G.; Chen, H.; Li, K.; Xiao, S. Porcine Epidemic Diarrhea Virus 3C-Like Protease Regulates Its Interferon Antagonism by Cleaving NEMO. J. Virol. 2016, 90, 2090–2101. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yuan, S.; Peng, Q.; Ding, Z.; Hao, W.; Peng, G.; Xiao, S.; Fang, L. Porcine Epidemic Diarrhea Virus nsp7 Inhibits Interferon-Induced JAK-STAT Signaling through Sequestering the Interaction between KPNA1 and STAT1. J. Virol. 2022, 96, e0040022. [Google Scholar] [CrossRef]
- Ding, Z.; Fang, L.; Jing, H.; Zeng, S.; Wang, D.; Liu, L.; Zhang, H.; Luo, R.; Chen, H.; Xiao, S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014, 88, 8936–8945. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Yang, R.; Lee, I.; Zhang, W.; Sun, J.; Wang, W.; Meng, X. Characterization of the SARS-CoV-2 E Protein: Sequence, Structure, Viroporin, and Inhibitors. Protein Sci. Publ. Protein Soc. 2021, 30, 1114–1130. [Google Scholar] [CrossRef]
- De Haan, C.; Vennema, H.; Rottier, P. Assembly of the coronavirus envelope: Homotypic interactions between the M proteins. J. Virol. 2000, 74, 4967–4978. [Google Scholar] [CrossRef]
- Vennema, H.; Godeke, G.; Rossen, J.; Voorhout, W.; Horzinek, M.; Opstelten, D.; Rottier, P. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996, 15, 2020–2028. [Google Scholar] [CrossRef]
- Den Boon, J.A.; Ahlquist, P. Organelle-Like Membrane Compartmentalization of Positive-Strand RNA Virus Replication Factories. Annu. Rev. Microbiol. 2010, 64, 241–256. [Google Scholar] [CrossRef]
- Krijnselocker, J.; Ericsson, M.; Rottier, P.J.M.; Griffiths, G. Characterization of the budding compartment of mouse hepatitis virus—Evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 1994, 124, 55–70. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C.J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe 2016, 20, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornillez-Ty, C.T.; Liao, L.; Yates III, J.R.; Kuhn, P.; Buchmeier, M.J. Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein 2 Interacts with a Host Protein Complex Involved in Mitochondrial Biogenesis and Intracellular Signaling. J. Virol. 2009, 83, 10314–10318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, G.; Sun, D.; Rajashankar, K.R.; Qian, Z.; Holmes, K.V.; Li, F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 10696–10701. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Prydz, K.; Saraste, J. The life cycle and enigmatic egress of coronaviruses. Mol. Microbiol. 2022, 117, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Arndt, A.L.; Larson, B.J.; Hogue, B.G. A Conserved Domain in the Coronavirus Membrane Protein Tail Is Important for Virus Assembly. J. Virol. 2010, 84, 11418–11428. [Google Scholar] [CrossRef] [Green Version]
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J.M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534. [Google Scholar] [CrossRef] [Green Version]
- Koshiba, T. Mitochondrial-mediated antiviral immunity. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Goping, I.S.; Millar, D.G.; Shore, G.C. Identification of the human mitochondrial protein import receptor, huMas20p—Complementation of Δmas20 in yeast. Febs Lett. 1995, 373, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ma, J.; Yoo, D. Inhibition of NF-κB activity by the porcine epidemic diarrhea virus nonstructural protein 1 for innate immune evasion. Virology 2017, 510, 111–126. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoo, D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016, 226, 128–141. [Google Scholar] [CrossRef]
- Davies, J.; Almasy, K.; McDonald, E.; Plate, L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect. Dis. 2020, 6, 3174–3189. [Google Scholar] [CrossRef]
- Hou, Y.; Meulia, T.; Gao, X.; Saif, L.J.; Wang, Q. Deletion of both the Tyrosine-Based Endocytosis Signal and the Endoplasmic Reticulum Retrieval Signal in the Cytoplasmic Tail of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Pigs. J. Virol. 2019, 93, e01758-18. [Google Scholar] [CrossRef] [Green Version]
- Cattin-Ortola, J.; Welch, L.G.; Maslen, S.L.; Papa, G.; James, L.C.; Munro, S. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Zou, D.; Xu, J.; Duan, X.; Xu, X.; Li, P.; Cheng, L.; Zheng, L.; Li, X.; Zhang, Y.; Wang, X.; et al. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019, 235, 209–219. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; Zhang, Q.; Huang, Y.; Dong, J.; Liang, Y.; Liu, H.-J.; Tong, D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013, 164, 212–221. [Google Scholar] [CrossRef]


| Name | Cellular/Subcellular Location |
|---|---|
| Nsp1 | Cyto. & Nuc. |
| Nsp2 | Cyto., Endoplasmic reticulum, Mitochondria |
| Nsp3 | Cyto., Endoplasmic reticulum |
| Nsp4 | Cyto., Endoplasmic reticulum |
| Nsp5 | Cyto. & Nuc. |
| Nsp6 | Cyto., Endoplasmic reticulum |
| Nsp7 | Cyto. & Nuc. |
| Nsp8 | Cyto. & Nuc. |
| Nsp9 | Cyto. & Nuc. |
| Nsp10 | Cyto. & Nuc. |
| Nsp12 | Cyto. & Nuc. |
| Nsp13 | Cyto. |
| Nsp14 | Cyto. & Nuc. |
| Nsp15 | Cyto. & Nuc. |
| Nsp16 | Cyto. & Nuc. |
| S1 | Cyto., Endoplasmic reticulum |
| S2 | Cyto., Endoplasmic reticulum (partially) |
| E | Cyto., Golgi apparatus, Mitochondria |
| M | Cyto., Golgi apparatus |
| N | Cyto., Endoplasmic reticulum (partially) |
| ORF3 | Cyto., Endoplasmic reticulum (partially) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Li, Z.; Bai, J.; Jiang, P.; Wang, X.; Liu, X. A Systemic Study of Subcellular Localization of Porcine Epidemic Diarrhea Virus Proteins. Pathogens 2022, 11, 1555. https://doi.org/10.3390/pathogens11121555
Zhu H, Li Z, Bai J, Jiang P, Wang X, Liu X. A Systemic Study of Subcellular Localization of Porcine Epidemic Diarrhea Virus Proteins. Pathogens. 2022; 11(12):1555. https://doi.org/10.3390/pathogens11121555
Chicago/Turabian StyleZhu, Huixin, Zitong Li, Juan Bai, Ping Jiang, Xianwei Wang, and Xing Liu. 2022. "A Systemic Study of Subcellular Localization of Porcine Epidemic Diarrhea Virus Proteins" Pathogens 11, no. 12: 1555. https://doi.org/10.3390/pathogens11121555






