Hexameric Aggregation Nucleation Core Sequences and Diversity of Pathogenic Tau Strains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Recombinant Truncated Tau Fragments Expression and Purification
2.3. Thioflavin-T Fluorescence Aggregation Kinetic Analysis
2.4. Protein Sequence Alignment
2.5. Structural Analysis of Tau Strains from Different Tauopathies
3. Results
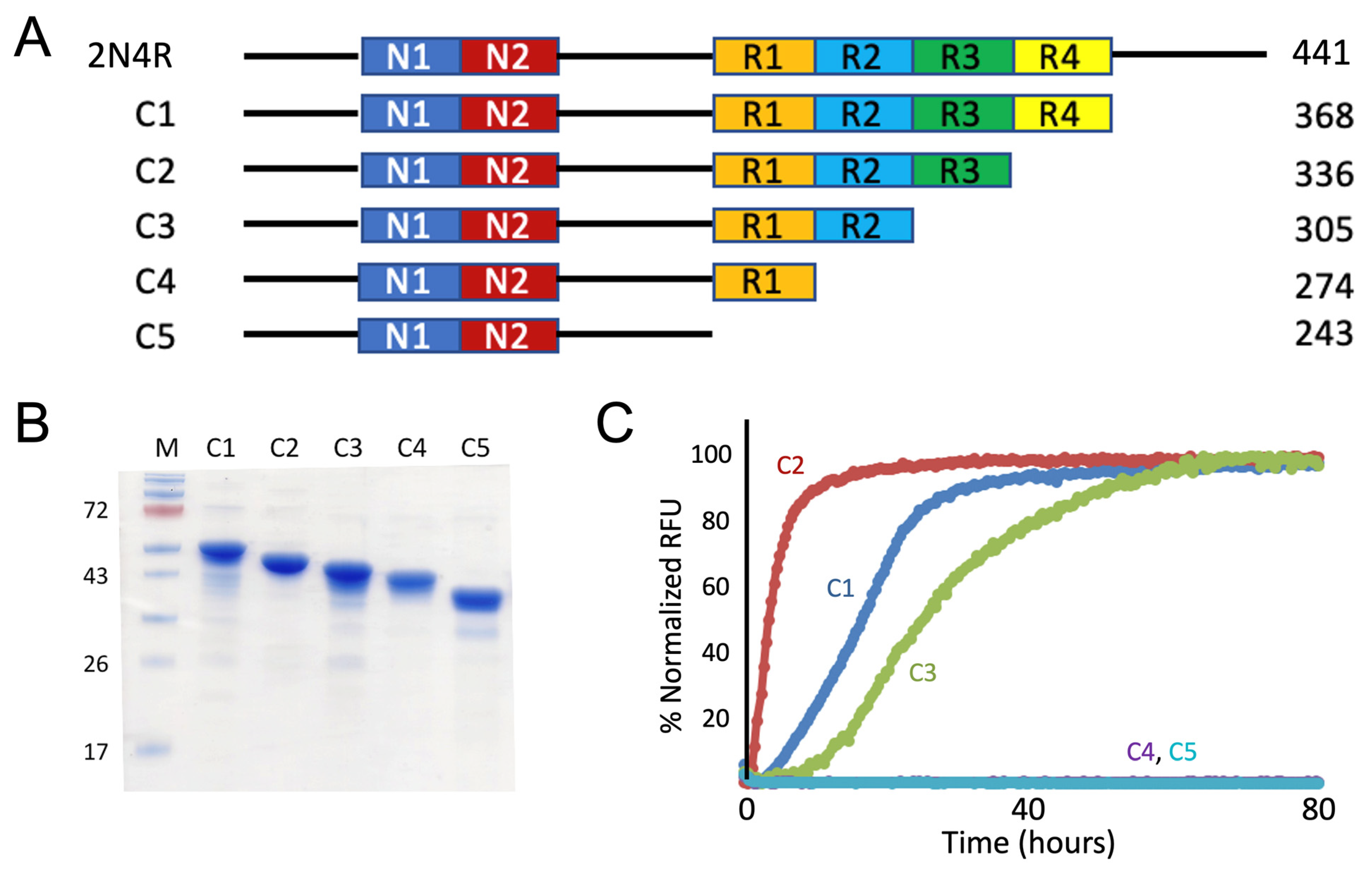
3.1. Truncation Mutagenesis of N-terminal & C-terminal Tau Sequences
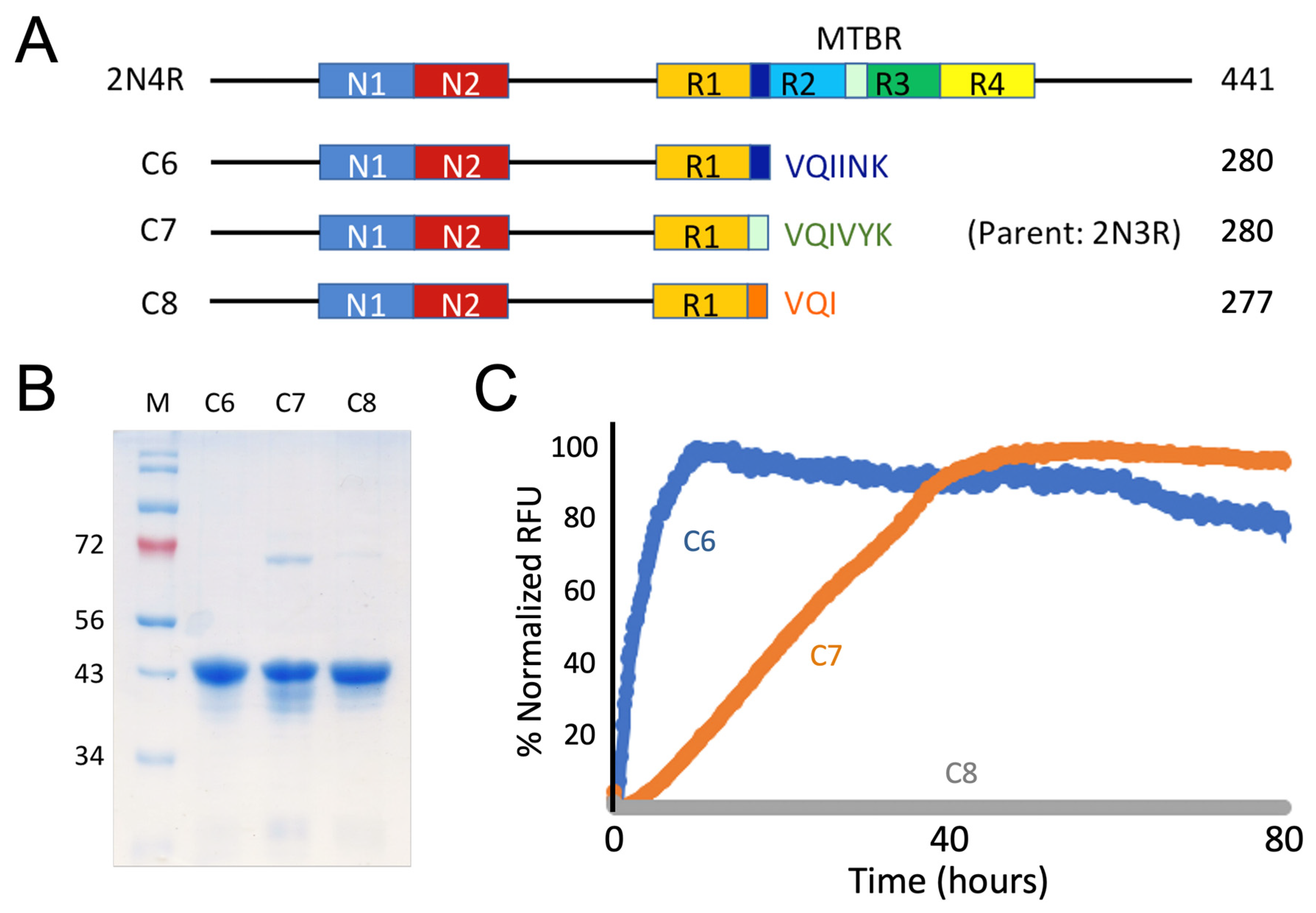
3.2. Recombinant Constructs with PHF6*, PHF6, and VQI Only Sequences
3.3. Hexameric Core Sequences in Diverse Pathogenic Tau Strains
3.4. Why Consensus VQI Sequence Is not Sufficient for Tau Aggregation?
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Braak, H.; Braak, E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef] [Green Version]
- Bloom, G.S. Amyloid-β and tau: The trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014, 71, 505–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spires-Jones, T.L.; Hyman, B.T. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 2014, 82, 756–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilcock, G.K.; Esiri, M.M. Plaques, tangles and dementia. A quantitative study. J. Neurol. Sci. 1982, 56, 343–356. [Google Scholar] [CrossRef]
- Nelson, P.T.; Alafuzoff, I.; Bigio, E.H.; Bouras, C.; Braak, H.; Cairns, N.J.; Castellani, R.J.; Crain, B.J.; Davies, P.; Tredici, K.D.; et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. 2012, 71, 362–381. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. Propagation of Tau aggregates. Mol. Brain 2017, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Arai, T.; Ikeda, K.; Akiyama, H.; Shikamoto, Y.; Tsuchiya, K.; Yagishita, S.; Beach, T.; Rogers, J.; Schwab, C.; McGeer, P.L. Distinct isoforms of tau aggregated in neurons and glial cells in brains of patients with Pick’s disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol. 2001, 101, 167–173. [Google Scholar] [CrossRef]
- Lee, V.M.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Cairns, N.J.; Crowther, R.A. Tau proteins of Alzheimer paired helical filaments: Abnormal phosphorylation of all six brain isoforms. Neuron 1992, 8, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, M.; de Silva, R.; Dickson, D.W.; Davies, P. Differential incorporation of tau isoforms in Alzheimer’s disease. J. Alzheimers Dis. 2008, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Togo, T.; Akiyama, H.; Iseki, E.; Uchikado, H.; Kondo, H.; Ikeda, K.; Tsuchiya, K.; de Silva, R.; Lees, A.; Kosaka, K. Immunohistochemical study of tau accumulation in early stages of Alzheimer-type neurofibrillary lesions. Acta Neuropathol. 2004, 107, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.D.; Kim, S.H.; Stein, T.D.; Pothast, M.J.; Nicks, R.; Meng, G.; Huber, B.R.; Mez, J.; Alosco, M.L.; Tripodis, Y.; et al. Evolution of neuronal and glial tau isoforms in chronic traumatic encephalopathy. Brain Pathol. 2020, 30, 913–925. [Google Scholar] [CrossRef]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef]
- Chung, D.C.; Roemer, S.; Petrucelli, L.; Dickson, D.W. Cellular and pathological heterogeneity of primary tauopathies. Mol. Neurodegener. 2021, 16, 57. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, W.; Yang, Y.; Murzin, A.G.; Falcon, B.; Kotecha, A.; van Beers, M.; Tarutani, A.; Kametani, F.; Garringer, H.J.; et al. Structure-based classification of tauopathies. Nature 2021, 598, 359–363. [Google Scholar] [CrossRef]
- Prusiner, S.B. Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 2013, 47, 601–623. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.L.; Lee, V.M. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011, 286, 15317–15331. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.L.; Narasimhan, S.; Changolkar, L.; He, Z.; Stieber, A.; Zhang, B.; Gathagan, R.J.; Iba, M.; McBride, J.D.; Trojanowski, J.Q.; et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016, 213, 2635–2654. [Google Scholar] [CrossRef]
- Swanson, E.; Breckenridge, L.; McMahon, L.; Som, S.; McConnell, L.; Bloom, G.S. Extracellular Tau Oligomers Induce Invation of Endogenous Tau into the Somatodendritic Compartment and Axonal Transport Dysfunction. J. Alzheimers Dis. 2017, 58, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, G.S.; Lee, V.M.Y.; Trojanowski, J.Q. Mechanisms of Cell-to-Cell Transmission of Pathological Tau: A Review. JAMA Neurol. 2019, 76, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Vaquer-Alicea, J.; Diamond, M.I. Propagation of Protein Aggregation in Neurodegenerative Diseases. Annu. Rev. Biochem. 2019, 88, 785–810. [Google Scholar] [CrossRef]
- Wang, Z.; Manca, M.; Foutz, A.; Martinez, M.C.; Raymond, G.J.; Race, B.; Orru, C.D.; Yuan, J.; Shen, P.; Li, B.; et al. Early Preclinical Detection of Prions in the Skin of Prion-Infected Animals. Nat. Commun. 2019, 10, 247. [Google Scholar] [CrossRef] [Green Version]
- Goedert, M. Tau proteinopathies and the prion concept. Prog. Mol. Biol. Transl. Sci. 2020, 175, 239–259. [Google Scholar] [PubMed]
- Kaufman, S.K.; Sanders, D.W.; Thomas, T.L.; Ruchinskas, A.J.; Vaquer-Alicea, J.; Sharma, A.M.; Miller, T.M.; Diamond, M.I. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability in vivo. Neuron 2016, 92, 796–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dujardin, S.; Bégard, S.; Caillierez, R.; Lachaud, C.; Carrier, S.; Lieger, S.; Gonzalez, J.A.; Deramecourt, V.; Déglon, N.; Maurage, C.A.; et al. Different tau species lead to heterogeneous tau pathology propagation and misfolding. Acta Neuropathol. Commun. 2018, 6, 132. [Google Scholar] [CrossRef] [Green Version]
- Vaquer-Alicea, J.; Diamond, M.I.; Joachimiak, L.A. Tau strains shape disease. Acta Neuropathol. 2021, 142, 57–71. [Google Scholar] [CrossRef]
- Xu, H.; O’Reilly, M.; Gibbons, G.S.; Changolkar, L.; McBride, J.D.; Riddle, D.M.; Zhang, B.; Stieber, A.; Nirschl, J.; Kim, S.J.; et al. In vitro amplification of pathogenic tau conserves disease-specific bioactive characteristics. Acta Neuropathol. 2021, 141, 193–215. [Google Scholar] [CrossRef]
- von Bergen, M.; Barghorn, S.; Müller, S.A.; Pickhardt, M.; Biernat, J.; Mandelkow, E.M.; Davies, P.; Aebi, U.; Mandelkow, E. The core of tau-paired helical filaments studied by scanning transmission electron microscopy and limited proteolysis. Biochemistry 2006, 45, 6446–6457. [Google Scholar] [CrossRef]
- von Bergen, M.; Friedhoff, P.; Biernat, J.; Heberle, J.; Mandelkow, E.M.; Mandelkow, E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. USA 2000, 97, 5129–5134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barghorn, S.; Biernat, J.; Mandelkow, E. Purification of recombinant tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol. Biol. 2005, 299, 35–51. [Google Scholar] [PubMed]
- Wu, L.; Wang, Z.; Lad, S.; Gilyazova, N.; Dougharty, D.T.; Marcus, M.; Henderson, F.; Ray, W.K.; Siedlak, S.; Li, J.; et al. Selective Detection of Misfolded Tau From Postmortem Alzheimer’s Disease Brains. Front. Aging Neurosci. 2022, 14, 945875. [Google Scholar] [CrossRef]
- Wu, L.; Velander, P.; Brown, A.M.; Wang, Y.; Liu, D.; Bevan, D.R.; Zhang, S.; Xu, B. Rosmarinic Acid Potently Detoxifies Amylin Amyloid and Ameliorates Diabetic Pathology in a Transgenic Rat Model of Type 2 Diabetes. ACS Pharmacol. Transl. Sci. 2021, 4, 1322–1337. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falcon, B.; Zivanov, J.; Zhang, W.; Murzin, A.G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Newell, K.L.; Ghetti, B.; Goedert, M.; et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 2019, 568, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zhang, W.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Ghetti, B.; Scheres, S.H.; Goedert, M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 2018, 561, 137–140. [Google Scholar] [CrossRef]
- Zhang, W.; Tarutani, A.; Newell, K.L.; Murzin, A.G.; Matsubara, T.; Falcon, B.; Vidal, R.; Garringer, H.J.; Shi, Y.; Ikeuchi, T.; et al. Novel tau filament fold in corticobasal degeneration. Nature 2020, 580, 283–287. [Google Scholar] [CrossRef]
- Velander, P.; Wu, L.; Hildreth, S.; Vogelaar, N.J.; Mukhopadhyay, B.; Zhang, S.; Helm, R.F.; Xu, B. Catechol-containing compounds are a broad class of amyloid inhibitors: Redox state is a key determinant of the inhibitory activities. Pharmacol. Res. 2022, 184, 106409. [Google Scholar] [CrossRef]
- Shahpasand-Kroner, H.; Portillo, J.; Lantz, C.; Seidler, P.M.; Sarafian, N.; Loo, J.A.; Bitan, G. Three-repeat and four-repeat tau isoforms form different oligomers. Protein Sci. 2022, 31, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Seidler, P.M.; Boyer, D.R.; Rodriguez, J.A.; Sawaya, M.R.; Cascio, D.; Murray, K.; Gonen, T.; Eisenberg, D.S. Structure-based inhibitors of tau aggregation. Nat. Chem. 2018, 10, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Saijo, E.; Ghetti, B.; Zanusso, G.; Oblak, A.; Furman, J.L.; Diamond, M.I.; Kraus, A.; Caughey, B. Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol. 2017, 133, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Kraus, A.; Saijo, E.; Metrick, M.A., 2nd; Newell, K.; Sigurdson, C.J.; Zanusso, G.; Ghetti, B.; Caughey, B. Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 2019, 137, 585–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metrick, M.A., 2nd; Ferreira, N.D.C.; Saijo, E.; Kraus, A.; Newell, K.; Zanusso, G.; Vendruscolo, M.; Ghetti, B.; Caughey, B. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol. Commun. 2020, 8, 22. [Google Scholar] [CrossRef]
- Arakhamia, T.; Lee, C.E.; Carlomagno, Y.; Kumar, M.; Duong, D.M.; Wesseling, H.; Kundinger, S.R.; Wang, K.; Williams, D.; DeTure, M.; et al. Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 2020, 180, 633–644. [Google Scholar] [CrossRef]
- Wesseling, H.; Mair, W.; Kumar, M.; Schlaffner, C.N.; Tang, S.; Beerepoot, P.; Fatou, B.; Guise, A.J.; Cheng, L.; Takeda, S.; et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer’s Disease. Cell 2020, 183, 1699–1713. [Google Scholar] [CrossRef]
- Wu, L.; Gilyzova, N.; Ervin, J.F.; Wang, S.J.; Xu, B. Site-Specific Phospho-Tau Aggregation-Based Biomarker Discovery for Alzheimer’s Diagnosis and Differentiation. ACS Chem. Neurosci. 2022, 13, 3281–3290. [Google Scholar] [CrossRef]
- Dujardin, S.; Commins, C.; Lathuiliere, A.; Beerepoot, P.; Fernandes, A.R.; Kamath, T.V.; De Los Santos, M.B.; Klickstein, N.; Corjuc, D.L.; Corjuc, B.T.; et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 2020, 26, 1256–1263. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Madhavan, S.S.; Tan, C.; Xu, B. Hexameric Aggregation Nucleation Core Sequences and Diversity of Pathogenic Tau Strains. Pathogens 2022, 11, 1559. https://doi.org/10.3390/pathogens11121559
Wu L, Madhavan SS, Tan C, Xu B. Hexameric Aggregation Nucleation Core Sequences and Diversity of Pathogenic Tau Strains. Pathogens. 2022; 11(12):1559. https://doi.org/10.3390/pathogens11121559
Chicago/Turabian StyleWu, Ling, Sidharth S. Madhavan, Christopher Tan, and Bin Xu. 2022. "Hexameric Aggregation Nucleation Core Sequences and Diversity of Pathogenic Tau Strains" Pathogens 11, no. 12: 1559. https://doi.org/10.3390/pathogens11121559





