Immunodominant B-Cell Linear Epitope on the VP1 P Domain of a Feline Norovirus Cat Model
Abstract
:1. Introduction
2. Results
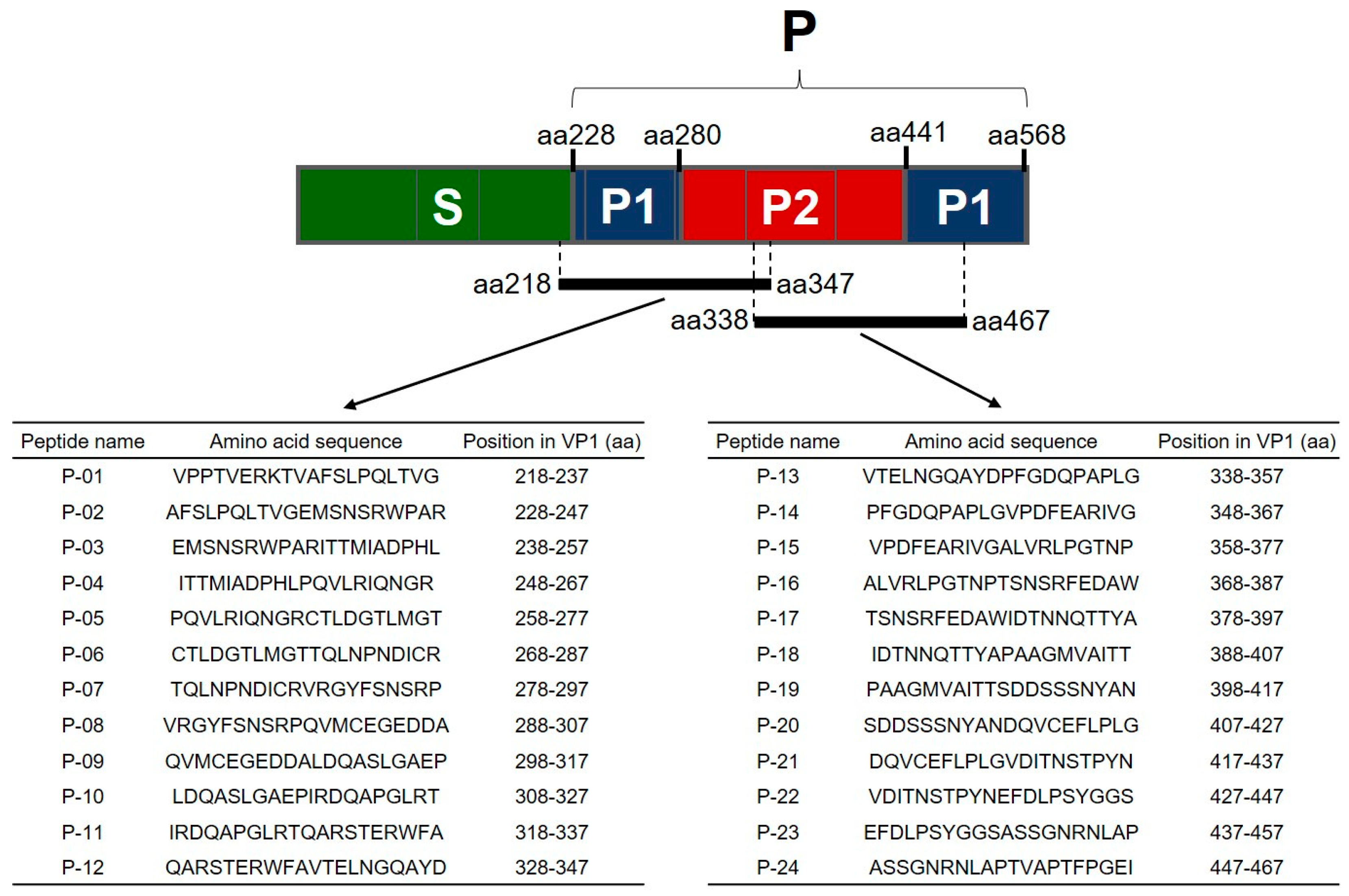
2.1. Mapping of the B-Cell Linear Epitopes in FNoV VP1
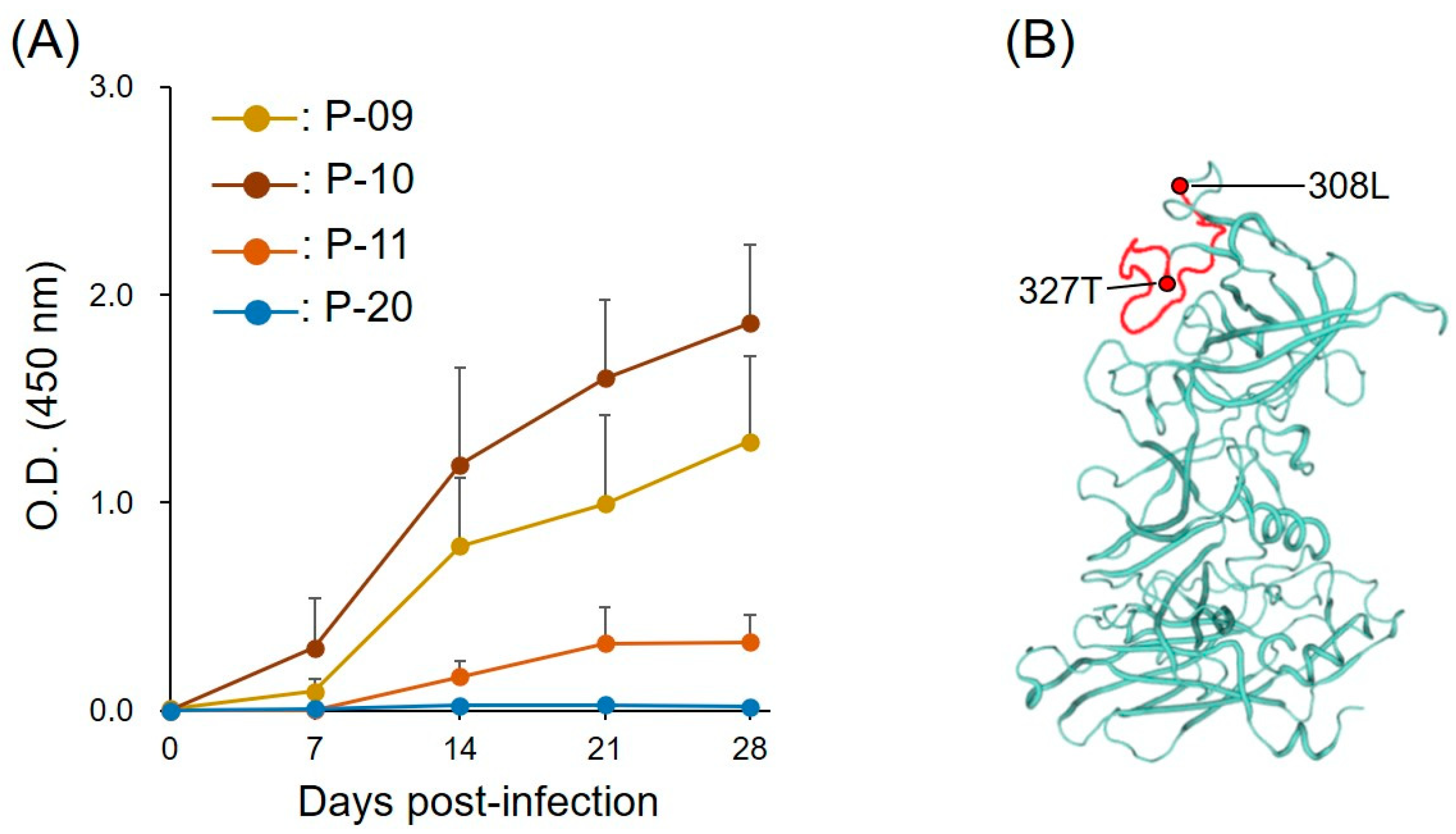
2.2. Kinetics of IgG Response to Peptides during Course of Infection
2.3. Location of B-Cell Linear Epitope in FNoV VP1
2.4. Viral Load, Clinical Symptoms, and Peptide-Specific Antibody Levels in Cats Infected with FNoV
3. Discussion
4. Materials and Methods
4.1. Peptides
4.2. Plasma Samples for Screening of Peptides
4.3. Peptide-Based ELISA
4.4. Homology Modelling of FNoV VP1
4.5. Ethics of Animal Experiments
4.6. Animal Infection
4.7. Statistical Analyse
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Glass, R.I.; Parashar, U.D.; Estes, M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009, 361, 1776–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar]
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global economic burden of norovirus gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, P.; Wang, Q.; Chen, N.; Dubovi, E.J.; Daniels, J.B.; Millward, L.M.; Buonavoglia, C.; Martella, V.; Saif, L.J. Discovery and genomic characterization of noroviruses from a gastroenteritis outbreak in domestic cats in the US. PLoS ONE 2012, 7, e32739. [Google Scholar]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Sarchese, V.; Cafiero, M.A.; Robetto, S.; Aste, G.; Lanave, G.; Marsilio, F.; Martella, V. A novel feline norovirus in diarrheic cats. Infect. Genet. Evol. 2016, 38, 132–137. [Google Scholar] [CrossRef]
- Mesquita, J.R.; Barclay, L.; Nascimento, M.S.J.; Vinjé, J. Novel norovirus in dogs with diarrhea. Emerg. Infect. Dis. 2010, 16, 980–982. [Google Scholar] [CrossRef]
- Ma, H.; Yue, H.; Luo, Y.; Li, S.; Tang, C. First detection of canine norovirus in dogs and a complete GVI. 2 genome in mainland China. Infect. Genet. Evol. 2021, 92, 104879. [Google Scholar] [CrossRef]
- Otto, P.H.; Clarke, I.N.; Lambden, P.R.; Salim, O.; Reetz, J.; Liebler-Tenorio, E.M. Infection of calves with bovine norovirus GIII. 1 strain Jena virus: An experimental model to study the pathogenesis of norovirus infection. J. Virol. 2011, 85, 12013–12021. [Google Scholar] [CrossRef] [Green Version]
- Vinjé, J.; Estes, M.K.; Esteves, P.; Green, K.Y.; Katayama, K.; Knowles, N.J.; L’Homme, Y.; Martella, V.; Vennema, H.; White, P.A.; et al. ICTV virus taxonomy profile: Caliciviridae. J. Gen. Virol. 2019, 100, 1469–1470. [Google Scholar] [CrossRef]
- Smith, H.Q.; Smith, T.J. The dynamic capsid structures of the noroviruses. Viruses 2019, 11, 235. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Jiang, X. Norovirus and its histo-blood group antigen receptors: An answer to a historical puzzle. Trends Microbiol. 2005, 13, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.W.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Cunha, J.B.; Elftman, M.D.; Kolawole, A.O. Animal models of norovirus infection. In Viral Gastroenteritis; Academic Press: Cambridge, MA, USA, 2016; pp. 397–422. [Google Scholar]
- Roth, A.N.; Helm, E.W.; Mirabelli, C.; Kirsche, E.; Smith, J.C.; Eurell, L.B.; Karst, S.M. Norovirus infection causes acute self-resolving diarrhea in wild-type neonatal mice. Nat. Commun. 2020, 11, 2968. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Marsilio, F. Feline Virome—A Review of Novel Enteric Viruses Detected in Cats. Viruses 2019, 11, 908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, T.A.; Eweida, A.E.; Sheweita, S.A. B-cell epitope mapping for the design of vaccines and effective diagnostics. Trials Vaccinol. 2016, 5, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Sivalingam, G.N.; Shepherd, A.J. An analysis of B-cell epitope discontinuity. Mol. Immunol. 2012, 51, 304–309. [Google Scholar] [CrossRef]
- Lucchese, G.; Stufano, A.; Trost, B.; Kusalik, A.; Kanduc, D. Peptidology: Short amino acid modules in cell biology and immunology. Amino Acids 2007, 33, 703–707. [Google Scholar] [CrossRef]
- Moeini, H.; Afridi, S.Q.; Donakonda, S.; Knolle, P.A.; Protzer, U.; Hoffmann, D. Linear B-Cell epitopes in human norovirus GII. 4 capsid protein elicit blockade antibodies. Vaccines 2021, 9, 52. [Google Scholar] [CrossRef]
- Parker, T.D.; Kitamoto, N.; Tanaka, T.; Hutson, A.M.; Estes, M.K. Identification of genogroup I and genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 2005, 79, 7402–7409. [Google Scholar] [CrossRef] [Green Version]
- Lindesmith, L.C.; Costantini, V.; Swanstrom, J.; Debbink, K.; Donaldson, E.F.; Vinjé, J.; Baric, R.S. Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J. Virol. 2013, 87, 2803–2813. [Google Scholar] [CrossRef] [Green Version]
- Mallory, M.L.; Lindesmith, L.C.; Graham, R.L.; Baric, R.S. GII.4 Human Norovirus: Surveying the Antigenic Landscape. Viruses 2019, 11, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanker, S.; Czakó, R.; Sapparapu, G.; Alvarado, G.; Viskovska, M.; Sankaran, B.; Atmar, R.L.; Crowe, J.E.; Estes, M.K., Jr.; Prasad, B.V. Structural basis for norovirus neutralization by an HBGA blocking human IgA antibody. Proc. Natl. Acad. Sci. USA 2016, 113, E5830–E5837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takano, T.; Hiramatsu, K.; Matsuyama, M.; Mutoh, K.; Matsumoto, Y.; Fukushima, T.; Doki, T.; Kusuhara, H.; Hohdatsu, T. Viral shedding and clinical status of feline-norovirus-infected cats after reinfection with the same strain. Arch. Virol. 2018, 163, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Thackray, L.B.; Virgin, H.W., IV. Murine norovirus: A model system to study norovirus biology and pathogenesis. J. Virol. 2006, 80, 5104–5112. [Google Scholar] [CrossRef] [Green Version]
- Hirneisen, K.A.; Kniel, K.E. Comparing human norovirus surrogates: Murine norovirus and Tulane virus. J. Food Prot. 2013, 76, 139–143. [Google Scholar] [CrossRef]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M.; et al. Median infectious dose of human norovirus GII. 4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J. Gen. Virol. 2013, 94, 2005–2016. [Google Scholar] [CrossRef]
- Souza, M.; Azevedo, M.D.S.P.D.; Jung, K.; Cheetham, S.; Saif, L.J. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II. 4-HS66 strain of human norovirus. J. Virol. 2008, 82, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Takano, T.; Kusuhara, H.; Kuroishi, A.; Takashina, M.; Doki, T.; Nishinaka, T.; Hohdatsu, T. Molecular characterization and pathogenicity of a genogroup GVI feline norovirus. Vet. Microbiol. 2015, 178, 201–207. [Google Scholar] [CrossRef]
- Thackray, L.B.; Wobus, C.E.; Chachu, K.A.; Liu, B.; Alegre, E.R.; Henderson, K.S.; Kelley, S.T.; Virgin, H.W., IV. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007, 81, 10460–10473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takano, T.; Watanabe, H.; Doki, T.; Kusuhara, H. Detection of feline norovirus using commercial real-time RT-PCR kit for the diagnosis of human norovirus infection. J. Vet. Med. Sci. 2021, 83, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Morioka, H.; Gomi, K.; Tomizawa, K.; Doki, T.; Hohdatsu, T. Screening and identification of T helper 1 and linear immunodominant antibody-binding epitopes in spike 1 domain and membrane protein of feline infectious peritonitis virus. Vaccine 2014, 32, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Qiao, N.; Wang, X.Y.; Liu, L. Temporal evolutionary dynamics of norovirus GII. 4 variants in China between 2004 and 2015. PLoS ONE 2016, 11, e0163166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Kahan, S.M.; Jia, Y.; Karst, S.M. Primary high-dose murine norovirus 1 infection fails to protect from secondary challenge with homologous virus. J. Virol. 2009, 83, 6963–6968. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takano, T.; Ryu, M.; Doki, T.; Kusuhara, H. Immunodominant B-Cell Linear Epitope on the VP1 P Domain of a Feline Norovirus Cat Model. Pathogens 2022, 11, 731. https://doi.org/10.3390/pathogens11070731
Takano T, Ryu M, Doki T, Kusuhara H. Immunodominant B-Cell Linear Epitope on the VP1 P Domain of a Feline Norovirus Cat Model. Pathogens. 2022; 11(7):731. https://doi.org/10.3390/pathogens11070731
Chicago/Turabian StyleTakano, Tomomi, Mizuki Ryu, Tomoyoshi Doki, and Hajime Kusuhara. 2022. "Immunodominant B-Cell Linear Epitope on the VP1 P Domain of a Feline Norovirus Cat Model" Pathogens 11, no. 7: 731. https://doi.org/10.3390/pathogens11070731





