High Rates of Multidrug-Resistant Escherichia coli in Great Cormorants (Phalacrocorax carbo) of the German Baltic and North Sea Coasts: Indication of Environmental Contamination and a Potential Public Health Risk
Abstract
:1. Introduction
2. Material and Methods
2.1. Sample Collection
2.2. E. coli Isolation and Identification
2.3. Resistance Phenotype
2.4. Resistance Genotype
2.5. Molecular Typing
2.6. Statistical Analysis
3. Results
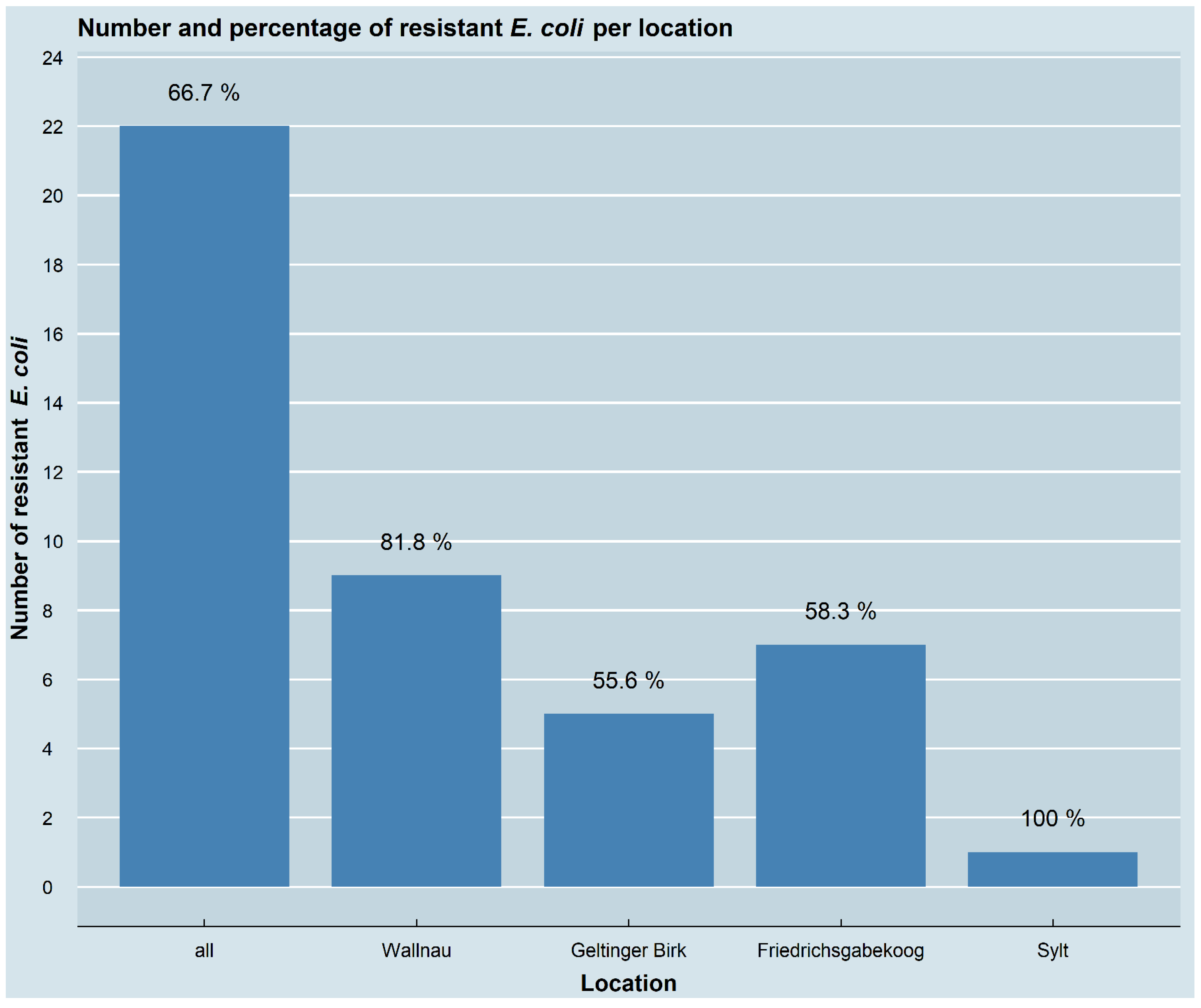
3.1. Occurrence of Antimicrobial-Resistant E. coli Isolates
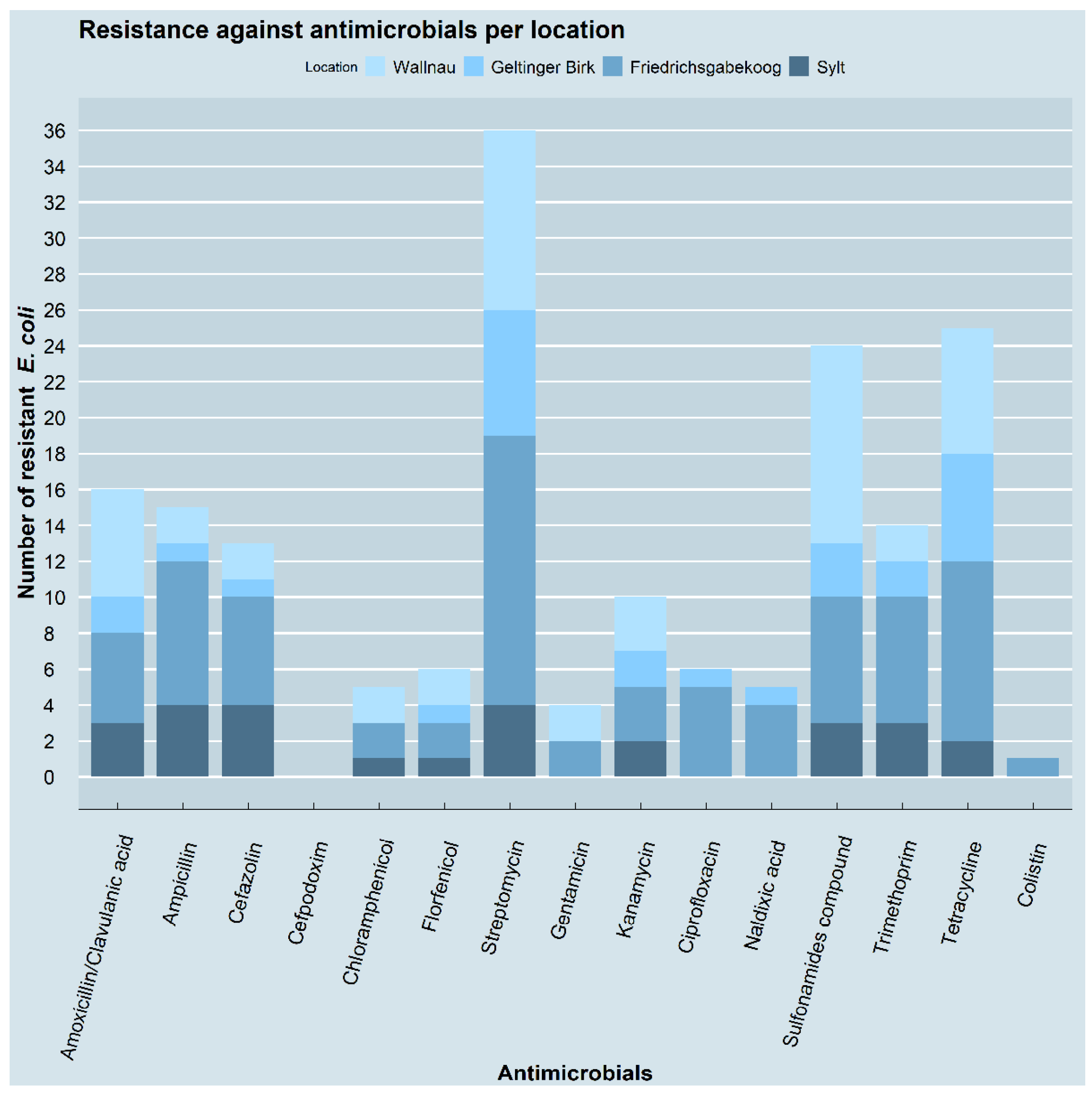
3.2. Resistance Pheno- and Genotypes
3.3. Molecular Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 5 May 2022).
- Acar, J.; Davies, J.; Buckley, M. Antibiotic Resistance: An Ecological Perspective on an Old Problem; American Society for Microbiology: Washington, DC, USA, 2009. [Google Scholar]
- Aminov, R.I. The Role of Antibiotics and Antibiotic Resistance in Nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Chattopadhyay, M.K.; Grossart, H.-P. The Multifaceted Roles of Antibiotics and Antibiotic Resistance in Nature. Front. Microbiol. 2013, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The Role of the Natural Environment in the Emergence of Antibiotic Resistance in Gram-Negative Bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Schijven, J.F.; Blaak, H.; Schets, F.M.; de Roda Husman, A.M. Fate of Extended-Spectrum β-Lactamase-Producing Escherichia coli from Faecal Sources in Surface Water and Probability of Human Exposure through Swimming. Environ. Sci. Technol. 2015, 49, 11825–11833. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, Á.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital Effluent: A Reservoir for Carbapenemase-Producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H.; et al. Comparable High Rates of Extended-Spectrum-Beta-Lactamase-Producing Escherichia coli in Birds of Prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, R.; Ternes, T.; Haberer, K.; Kratz, K.-L. Occurrence of Antibiotics in the Aquatic Environment. Sci. Total Environ. 1999, 225, 109–118. [Google Scholar] [CrossRef]
- Boxall, A.B.A.; Johnson, P.; Smith, E.J.; Sinclair, C.J.; Stutt, E.; Levy, L.S. Uptake of Veterinary Medicines from Soils into Plants. J. Agric. Food Chem. 2006, 54, 2288–2297. [Google Scholar] [CrossRef]
- Milić, N.; Milanović, M.; Letić, N.G.; Sekulić, M.T.; Radonić, J.; Mihajlović, I.; Miloradov, M.V. Occurrence of Antibiotics as Emerging Contaminant Substances in Aquatic Environment. Int. J. Environ. Health Res. 2013, 23, 296–310. [Google Scholar] [CrossRef]
- Monteiro, S.C.; Boxall, A.B.A. Occurrence and fate of human pharmaceuticals in the environment. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2010; Volume 202, pp. 53–154. ISBN 9781441911568. [Google Scholar]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential Impact of Antimicrobial Resistance in Wildlife, Environment and Human Health. Front. Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Halpern, B.S.; Walbridge, S.; Selkoe, K.A.; Kappel, C.V.; Micheli, F.; D’Agrosa, C.; Bruno, J.F.; Casey, K.S.; Ebert, C.; Fox, H.E.; et al. A Global Map of Human Impact on Marine Ecosystems. Science 2008, 319, 948–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooban, B.; Fitzhenry, K.; Cahill, N.; Joyce, A.; O’Connor, L.; Bray, J.E.; Brisse, S.; Passet, V.; Abbas Syed, R.; Cormican, M.; et al. A Point Prevalence Survey of Antibiotic Resistance in the Irish Environment, 2018–2019. Environ. Int. 2021, 152, 106466. [Google Scholar] [CrossRef] [PubMed]
- Mahon, B.M.; Brehony, C.; McGrath, E.; Killeen, J.; Cormican, M.; Hickey, P.; Keane, S.; Hanahoe, B.; Dolan, A.; Morris, D. Indistinguishable NDM-Producing Escherichia coli Isolated from Recreational Waters, Sewage, and a Clinical Specimen in Ireland, 2016 to 2017. Eurosurveillance 2017, 22, 30513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic Resistance in Recreational Waters: State of the Science. Int. J. Environ. Res. Public Health 2020, 17, 8034. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.L.; Joyce, A.; Duane, S.; Fitzhenry, K.; Hooban, B.; Burke, L.P.; Morris, D. Evaluating the Potential for Exposure to Organisms of Public Health Concern in Naturally Occurring Bathing Waters in Europe: A Scoping Review. Water Res. 2021, 206, 117711. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and Fate of Antibiotic Resistant Bacteria in Wastewater Treatment Plants: A Review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Kotlarska, E.; Łuczkiewicz, A.; Pisowacka, M.; Burzyński, A. Antibiotic Resistance and Prevalence of Class 1 and 2 Integrons in Escherichia coli Isolated from Two Wastewater Treatment Plants, and Their Receiving Waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 2015, 22, 2018–2030. [Google Scholar] [CrossRef] [Green Version]
- Garcia, S.; Wade, B.; Bauer, C.; Craig, C.; Nakaoka, K.; Lorowitz, W. The Effect of Wastewater Treatment on Antibiotic Resistance in Escherichia coli and Enterococcus sp. Water Environ. Res. 2007, 79, 2387–2395. [Google Scholar] [CrossRef]
- Lachmayr, K.L.; Kerkhof, L.J.; DiRienzo, A.G.; Cavanaugh, C.M.; Ford, T.E. Quantifying Nonspecific TEM β-Lactamase (BlaTEM) Genes in a Wastewater Stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Nagulapally, S.R.; Ahmad, A.; Henry, A.; Marchin, G.L.; Zurek, L.; Bhandari, A. Occurrence of Ciprofloxacin-, Trimethoprim-Sulfamethoxazole-, and Vancomycin-Resistant Bacteria in a Municipal Wastewater Treatment Plant. Water Environ. Res. 2009, 81, 82–90. [Google Scholar] [CrossRef]
- Zhang, Y.; Marrs, C.F.; Simon, C.; Xi, C. Wastewater Treatment Contributes to Selective Increase of Antibiotic Resistance among Acinetobacter spp. Sci. Total Environ. 2009, 407, 3702–3706. [Google Scholar] [CrossRef] [PubMed]
- Quach-Cu, J.; Herrera-Lynch, B.; Marciniak, C.; Adams, S.; Simmerman, A.; Reinke, R. The Effect of Primary, Secondary, and Tertiary Wastewater Treatment Processes on Antibiotic Resistance Gene (ARG) Concentrations in Solid and Dissolved Wastewater Fractions. Water 2018, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Börjesson, S.; Matussek, A.; Melin, S.; Löfgren, S.; Lindgren, P.E. Methicillin-Resistant Staphylococcus aureus (MRSA) in Municipal Wastewater: An Uncharted Threat? J. Appl. Microbiol. 2010, 108, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [Green Version]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [Green Version]
- Cooper, R.M.; Tsimring, L.; Hasty, J. Inter-Species Population Dynamics Enhance Microbial Horizontal Gene Transfer and Spread of Antibiotic Resistance. Elife 2017, 6, e25950. [Google Scholar] [CrossRef]
- Hooban, B.; Joyce, A.; Fitzhenry, K.; Chique, C.; Morris, D. The Role of the Natural Aquatic Environment in the Dissemination of Extended Spectrum Beta-Lactamase and Carbapenemase Encoding Genes: A Scoping Review. Water Res. 2020, 180, 115880. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; Hill, S. Occurrence of Antibiotic Resistance in Escherichia coli from Surface Waters and Fecal Pollution Sources near Hamilton, Ontario. Can. J. Microbiol. 2005, 51, 501–505. [Google Scholar] [CrossRef]
- Yuan, Y.; Liang, B.; Jiang, B.; Zhu, L.; Wang, T.; Li, Y.; Liu, J.; Guo, X.; Ji, X.; Sun, Y. Migratory Wild Birds Carrying Multidrug-Resistant Escherichia coli as Potential Transmitters of Antimicrobial Resistance in China. PLoS ONE 2021, 16, e0261444. [Google Scholar] [CrossRef]
- Riedel, S.; Boire, N.; Carson, K.A.; Vadlamudi, A.; Khuvis, J.; Vadlamudi, V.; Atukorale, V.; Riedel, V.A.A.; Parrish, N.M. A Survey of Antimicrobial Resistance in Enterobacteriaceae Isolated from the Chesapeake Bay and Adjacent Upper Tributaries. MicrobiologyOpen 2019, 8, e00839. [Google Scholar] [CrossRef] [Green Version]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Szmolka, A.; Nagy, B. Multidrug Resistant Commensal Escherichia coli in Animals and Its Impact for Public Health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 979–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Elsas, J.D.; Semenov, A.V.; Costa, R.; Trevors, J.T. Survival of Escherichia coli in the Environment: Fundamental and Public Health Aspects. ISME J. 2011, 5, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Marcelino, V.R.; Wille, M.; Hurt, A.C.; González-Acuña, D.; Klaassen, M.; Schlub, T.E.; Eden, J.-S.; Shi, M.; Iredell, J.R.; Sorrell, T.C.; et al. Meta-Transcriptomics Reveals a Diverse Antibiotic Resistance Gene Pool in Avian Microbiomes. BMC Biol. 2019, 17, 31. [Google Scholar] [CrossRef] [Green Version]
- Guenther, S.; Ewers, C.; Wieler, L.H. Extended-Spectrum Beta-Lactamases Producing E. coli in Wildlife, yet Another Form of Environmental Pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.M.; Kolenda, R.; Schierack, P.; Weinreich, J.; Rödiger, S.; Schierack, J.; Stubbe, M.; Lkhagvasuren, D.; Guenther, S.; Schaufler, K. Investigation of Commensal Escherichia coli Populations of Cormorant Hatchlings in the Absence of Anthropogenic Impacts in Remote Areas of West Mongolia. Microorganisms 2021, 9, 372. [Google Scholar] [CrossRef]
- Lin, Y.; Dong, X.; Sun, R.; Wu, J.; Tian, L.; Rao, D.; Zhang, L.; Yang, K. Migratory Birds-One Major Source of Environmental Antibiotic Resistance around Qinghai Lake, China. Sci. Total Environ. 2020, 739, 139758. [Google Scholar] [CrossRef] [PubMed]
- Jamborova, I.; Johnston, B.D.; Papousek, I.; Kachlikova, K.; Micenkova, L.; Clabots, C.; Skalova, A.; Chudejova, K.; Dolejska, M.; Literak, I.; et al. Extensive Genetic Commonality among Wildlife, Wastewater, Community, and Nosocomial Isolates of Escherichia coli Sequence Type 131 (H 30R1 and H 30Rx Subclones) That Carry Bla CTX-M-27 or Bla CTX-M-15. Antimicrob. Agents Chemother. 2018, 62, e00519-18. [Google Scholar] [CrossRef] [Green Version]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.D.; Wieler, L.H.; Ewers, C. Antimicrobial Resistance Profiles of Escherichia coli from Common European Wild Bird Species. Vet. Microbiol. 2010, 144, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Blyton, M.D.J.; Pi, H.; Vangchhia, B.; Abraham, S.; Trott, D.J.; Johnson, J.R.; Gordon, D.M. Genetic Structure and Antimicrobial Resistance of Escherichia coli and Cryptic Clades in Birds with Diverse Human Associations. Appl. Environ. Microbiol. 2015, 81, 5123–5133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczkowski, M.; Krawiec, M.; Voslamber, B.; Książczyk, M.; Płoskońska-Bugla, G.; Wieliczko, A. Virulence Genes and the Antimicrobial Susceptibility of Escherichia coli, Isolated from Wild Waterbirds, in The Netherlands and Poland. Vector-Borne Zoonotic Dis. 2016, 16, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Skarżyńska, M.; Zając, M.; Bomba, A.; Bocian, Ł.; Kozdruń, W.; Polak, M.; Wiącek, J.; Wasyl, D. Antimicrobial Resistance Glides in the Sky—Free-Living Birds as a Reservoir of Resistant Escherichia coli With Zoonotic Potential. Front. Microbiol. 2021, 12, 656223. [Google Scholar] [CrossRef]
- Gaye-Siessegger, J. The Great Cormorant (Phalacrocorax Carbo) at Lower Lake Constance/Germany: Dietary Composition and Impact on Commercial Fisheries. Knowl. Manag. Aquat. Ecosyst. 2014, 414, 4. [Google Scholar] [CrossRef] [Green Version]
- Gagliardi, A.; Preatoni, D.G.; Wauters, L.A.; Martinoli, A. Selective Predators or Choosy Fishermen? Relation between Fish Harvest, Prey Availability and Great Cormorant (Phalacrocorax carbo sinensis) Diet. Ital. J. Zool. 2015, 82, 544–555. [Google Scholar] [CrossRef] [Green Version]
- Lehikoinen, A.; Heikinheimo, O.; Lappalainen, A. Temporal Changes in the Diet of Great Cormorant (Phalacrocorax carbo sinensis) on the Southern Coast of Finland—Comparison with Available Fish Data. Boreal Environ. Res. 2011, 16, 61–70. [Google Scholar]
- Dias, E.; Morais, P.; Leopold, M.; Campos, J.; Antunes, C. Natural Born Indicators: Great Cormorant Phalacrocorax carbo (Aves: Phalacrocoracidae) as Monitors of River Discharge Influence on Estuarine Ichthyofauna. J. Sea Res. 2012, 73, 101–108. [Google Scholar] [CrossRef]
- Tausova, D.; Dolejska, M.; Cizek, A.; Hanusova, L.; Hrusakova, J.; Svoboda, O.; Camlik, G.; Literak, I. Escherichia coli with Extended-Spectrum -Lactamase and Plasmid-Mediated Quinolone Resistance Genes in Great Cormorants and Mallards in Central Europe. J. Antimicrob. Chemother. 2012, 67, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Odoi, J.O.; Sugiyama, M.; Kitamura, Y.; Sudo, A.; Omatsu, T.; Asai, T. Prevalence of Antimicrobial Resistance in Bacteria Isolated from Great Cormorants (Phalacrocorax carbo hanedae) in Japan. J. Vet. Med. Sci. 2021, 83, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Stephan, R.; Hächler, H. Higher-Generation Cephalosporin-Resistant Escherichia coli in Feral Birds in Switzerland. Int. J. Antimicrob. Agents 2013, 41, 296–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bregnballe, T.; Frederiksen, M.; Gregersen, J. Seasonal Distribution and Timing of Migration of Cormorants Phalacrocorax carbo sinensis Breeding in Denmark. Bird Study 1997, 44, 257–276. [Google Scholar] [CrossRef]
- Bregnballe, T.; Frederiksen, M.; Gregersen, J. Effects of Distance to Wintering Area on Arrival Date and Breeding Performance in Great Cormorants Phalacrocorax carbo. Ardea 2006, 94, 619–630. [Google Scholar]
- Doumith, M.; Day, M.J.; Hope, R.; Wain, J.; Woodford, N. Improved Multiplex PCR Strategy for Rapid Assignment of the Four Major Escherichia coli Phylogenetic Groups. J. Clin. Microbiol. 2012, 50, 3108–3110. [Google Scholar] [CrossRef] [Green Version]
- CLSI Standard VET01; CLSI Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 5th ed. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2018.
- CLSI Supplement VET01S; CLSI Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 5th ed. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2020.
- CLSI Supplement M100; CLSI Performance Standards for Antimicrobial Susceptibility Testing. 32nd ed. Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2022.
- Müller, A.; Jansen, W.; Grabowski, N.T.; Kehrenberg, C. Characterization of Salmonella enterica Serovars Recovered from Meat Products Legally and Illegally Imported into the EU Reveals the Presence of Multiresistant and AmpC-Producing Isolates. Gut Pathog. 2018, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Randall, L.P.; Cooles, S.W.; Piddock, L.J.V.; Woodward, M.J. Effect of Triclosan or a Phenolic Farm Disinfectant on the Selection of Antibiotic-Resistant Salmonella enterica. J. Antimicrob. Chemother. 2004, 54, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Kehrenberg, C.; Schwarz, S. Occurrence and Linkage of Genes Coding for Resistance to Sulfonamides, Streptomycin and Chloramphenicol in Bacteria of the Genera Pasteurella and Mannheimia. FEMS Microbiol. Lett. 2001, 205, 283–290. [Google Scholar] [CrossRef]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial Resistance and Resistance Gene Determinants in Clinical Escherichia coli from Different Animal Species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Agersø, Y.; Ahrens, P.; Jørgensen, J.C.Ø.; Madsen, M.; Jensen, L.B. Antimicrobial Susceptibility and Presence of Resistance Genes in Staphylococci from Poultry. Vet. Microbiol. 2000, 74, 353–364. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, Mcr-1, Mcr-2, Mcr-3, Mcr-4 and Mcr-5 for Surveillance Purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a Novel Mcr-6 to Mcr-9 Multiplex PCR and Assessment of Mcr-1 to Mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates from Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef] [Green Version]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of Minimal Predictors of Avian Pathogenic Escherichia coli Virulence for Use as a Rapid Diagnostic Tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribot, E.M.; Fair, M.A.; Gautom, R.; Cameron, D.N.; Hunter, S.B.; Swaminathan, B.; Barrett, T.J. Standardization of Pulsed-Field Gel Electrophoresis Protocols for the Subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 2006, 3, 59–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobbin, G.; Hariharan, H.; Daoust, P.-Y.; Hariharan, S.; Heaney, S.; Coles, M.; Price, L.; Anne Muckle, C. Bacterial Flora of Free-Living Double-Crested Cormorant (Phalacrocorax auritus) Chicks on Prince Edward Island, Canada, with Reference to Enteric Bacteria and Antibiotic Resistance. Comp. Immunol. Microbiol. Infect. Dis. 2005, 28, 71–82. [Google Scholar] [CrossRef]
- Nowaczek, A.; Dec, M.; Stępień-Pyśniak, D.; Urban-Chmiel, R.; Marek, A.; Różański, P. Antibiotic Resistance and Virulence Profiles of Escherichia coli Strains Isolated from Wild Birds in Poland. Pathogens 2021, 10, 1059. [Google Scholar] [CrossRef] [PubMed]
- Radhouani, H.; Poeta, P.; Igrejas, G.; Gonçalves, A.; Vinué, L.; Torres, C. Antimicrobial Resistance and Phylogenetic Groups in Isolates of Escherichia coli from Seagulls at the Berlengas Nature Reserve. Vet. Rec. 2009, 165, 138–142. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Hernandez, J.; Stedt, J.; Waldenström, J.; Olsen, B.; Drobni, M. Extended-Spectrum β-Lactamases in Escherichia coli and Klebsiella pneumoniae in Gulls, Alaska, USA. Emerg. Infect. Dis. 2014, 20, 897–899. [Google Scholar] [CrossRef]
- Sacristán, C.; Esperón, F.; Herrera-León, S.; Iglesias, I.; Neves, E.; Nogal, V.; Muñoz, M.J.; de la Torre, A. Virulence Genes, Antibiotic Resistance and Integrons in Escherichia coli Strains Isolated from Synanthropic Birds from Spain. Avian Pathol. 2014, 43, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Stedt, J.; Bonnedahl, J.; Hernandez, J.; McMahon, B.J.; Hasan, B.; Olsen, B.; Drobni, M.; Waldenström, J. Antibiotic Resistance Patterns in Escherichia coli from Gulls in Nine European Countries. Infect. Ecol. Epidemiol. 2014, 4, 21565. [Google Scholar] [CrossRef] [Green Version]
- Dolejska, M.; Cizek, A.; Literak, I. High Prevalence of Antimicrobial-Resistant Genes and Integrons in Escherichia coli Isolates from Black-Headed Gulls in the Czech Republic. J. Appl. Microbiol. 2007, 103, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Literak, I.; Dolejska, M.; Janoszowska, D.; Hrusakova, J.; Meissner, W.; Rzyska, H.; Bzoma, S.; Cizek, A. Antibiotic-Resistant Escherichia coli Bacteria, Including Strains with Genes Encoding the Extended-Spectrum Beta-Lactamase and QnrS, in Waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 2010, 76, 8126–8134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hathcock, T.; Poudel, A.; Kang, Y.; Butaye, P.; Raiford, D.; Mobley, T.; Wang, C.; Bellah, J. Multidrug-Resistant Escherichia coli and Tetracycline-Resistant Enterococcus faecalis in Wild Raptors of Alabama and Georgia, USA. J. Wildl. Dis. 2019, 55, 482. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Hernandez, J.; Tyrlöv, V.; Uher-Koch, B.D.; Schmutz, J.A.; Atterby, C.; Järhult, J.D.; Bonnedahl, J. Antibiotic-Resistant Escherichia coli in Migratory Birds Inhabiting Remote Alaska. Ecohealth 2018, 15, 72–81. [Google Scholar] [CrossRef]
- Cole, D.; Drum, D.J.V.; Stallknecht, D.E.; Lee, M.D.; Ayers, S.; Sobsey, M.; Maurert, J.J. Free-Living Canada Geese and Antimicrobiai Resistance. Emerg. Infect. Dis. 2005, 11, 935–938. [Google Scholar] [CrossRef]
- Atterby, C.; Ramey, A.M.; Hall, G.G.; Järhult, J.; Börjesson, S.; Bonnedahl, J. Increased Prevalence of Antibiotic-Resistant E. coli in Gulls Sampled in Southcentral Alaska Is Associated with Urban Environments. Infect. Ecol. Epidemiol. 2016, 6, 32334. [Google Scholar] [CrossRef]
- Skurnik, D. Effect of Human Vicinity on Antimicrobial Resistance and Integrons in Animal Faecal Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 1215–1219. [Google Scholar] [CrossRef]
- Kaba, H.E.J.; Kuhlmann, E.; Scheithauer, S. Thinking Outside the Box: Association of Antimicrobial Resistance with Climate Warming in Europe—A 30 Country Observational Study. Int. J. Hyg. Environ. Health 2020, 223, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Schleswig-Holstein. Schleswig-Holstein—Zahlen Zur Bevölkerung. Available online: https://www.schleswig-holstein.de/DE/LandLeute/ZahlenFakten/_documents/bevoelkerung.html (accessed on 27 May 2022).
- World Health Organization. WHO Access, Watch, Reserve (AWaRe) Classification of Antibiotics for Evaluation and Monitoring of Use. 2021. Geneva: World Health Organization; (WHO/MHP/HPS/EML/2021.04); License: CC BY-NC-SA 3.0 IGO; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Bundesamt für Verbraucherschutz und Lebensmittelsicherheit. Paul-Ehrlich-Gesellschaft für Chemotherapie e.V. GERMAP 2015—Bericht Über Den Antibiotikaverbrauch Und Die Verbreitung von Antibiotikaresistenzen in Der Human- Und Veterinärmedizin in Deutschland; Antiinfectives Intelligence: Rheinbach, Germany, 2016. [Google Scholar]
- Guenther, S.; Grobbel, M.; Beutlich, J.; Bethe, A.; Friedrich, N.D.; Goedecke, A.; Lübke-Becker, A.; Guerra, B.; Wieler, L.H.; Ewers, C. CTX-M-15-Type Extended-Spectrum Beta-Lactamases-Producing Escherichia coli from Wild Birds in Germany. Environ. Microbiol. Rep. 2010, 2, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of Drug Resistance: Quinolone Resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef] [Green Version]
- Araújo, B.F.; de Campos, P.A.; Royer, S.; Ferreira, M.L.; Gonçalves, I.R.; da Fonseca Batistão, D.W.; Resende, D.S.; de Brito, C.S.; Gontijo-Filho, P.P.; Ribas, R.M. High Frequency of the Combined Presence of QRDR Mutations and PMQR Determinants in Multidrug-Resistant Klebsiella pneumoniae and Escherichia coli Isolates from Nosocomial and Community-Acquired Infections. J. Med. Microbiol. 2017, 66, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Kotb, D.N.; Mahdy, W.K.; Mahmoud, M.S.; Khairy, R.M.M. Impact of Co-Existence of PMQR Genes and QRDR Mutations on Fluoroquinolones Resistance in Enterobacteriaceae Strains Isolated from Community and Hospital Acquired UTIs. BMC Infect. Dis. 2019, 19, 979. [Google Scholar] [CrossRef]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Samadi Kafil, H. Molecular Mechanisms Related to Colistin Resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.; Martinez, J. Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Webber, M.A. The Importance of Efflux Pumps in Bacterial Antibiotic Resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef]
- Bingen, E.; Picard, B.; Brahimi, N.; Mathy, S.; Desjardins, P.; Elion, J.; Denamur, E. Phylogenetic Analysis of Escherichia coli Strains Causing Neonatal Meningitis Suggests Horizontal Gene Transfer from a Predominant Pool of Highly Virulent B2 Group Strains. J. Infect. Dis. 1998, 177, 642–650. [Google Scholar] [CrossRef] [Green Version]
- Boyd, E.F.; Hartl, D.L. Chromosomal Regions Specific to Pathogenic Isolates of Escherichia coli Have a Phylogenetically Clustered Distribution. J. Bacteriol. 1998, 180, 1159–1165. [Google Scholar] [CrossRef] [Green Version]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The Link between Phylogeny and Virulence in Escherichia coli Extraintestinal Infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Gall, T.; Clermont, O.; Gouriou, S.; Picard, B.; Nassif, X.; Denamur, E.; Tenaillon, O. Extraintestinal Virulence Is a Coincidental By-Product of Commensalism in B2 Phylogenetic Group Escherichia coli Strains. Mol. Biol. Evol. 2007, 24, 2373–2384. [Google Scholar] [CrossRef] [Green Version]
- Radhouani, H.; Poeta, P.; Gonçalves, A.; Pacheco, R.; Sargo, R.; Igrejas, G. Wild Birds as Biological Indicators of Environmental Pollution: Antimicrobial Resistance Patterns of Escherichia coli and Enterococci Isolated from Common Buzzards (Buteo buteo). J. Med. Microbiol. 2012, 61, 837–843. [Google Scholar] [CrossRef]
- Ishii, S.; Meyer, K.P.; Sadowsky, M.J. Relationship between Phylogenetic Groups, Genotypic Clusters, and Virulence Gene Profiles of Escherichia coli Strains from Diverse Human and Animal Sources. Appl. Environ. Microbiol. 2007, 73, 5703–5710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, J.; Hohn, C.; Hornor, S.; Frana, M.; Denver, M.; Joerger, R. Genotyping of Escherichia coli from Environmental and Animal Samples. J. Microbiol. Methods 2007, 70, 227–235. [Google Scholar] [CrossRef] [PubMed]



| Resistance Gene | Wall | Gelt | Frie | Sylt | Total |
|---|---|---|---|---|---|
| blaTEM | 2 | 1 | 8 | 4 | 15 |
| strA | 2 | 2 | 13 | 1 | 18 |
| strB | 2 | 2 | 13 | 1 | 18 |
| aadA1 | 8 | 0 | 2 | 1 | 11 |
| aac(3)-II | 0 | 0 | 1 | 0 | 1 |
| ant(2“)-I | 2 | 0 | 0 | 0 | 2 |
| floR | 2 | 0 | 2 | 1 | 5 |
| tet(A) | 2 | 6 | 6 | 2 | 16 |
| tet(B) | 5 | 0 | 5 | 0 | 10 |
| tet(D) | 0 | 1 | 0 | 0 | 1 |
| sul1 | 8 | 3 | 3 | 0 | 14 |
| sul2 | 9 | 2 | 7 | 2 | 20 |
| sul3 | 0 | 0 | 0 | 1 | 1 |
| dfrA5/A14 | 2 | 0 | 2 | 2 | 6 |
| dfrA7/A17 | 0 | 2 | 1 | 0 | 3 |
| dfrA1/15/16 | 0 | 0 | 4 | 1 | 5 |
| qnrS | 0 | 0 | 2 | 0 | 2 |
| Phylogenetic Group | A | B1 | B2 | D |
|---|---|---|---|---|
| Wallnau | 0 | 8 (53.3%) | 2 (13.3) | 5 (33.3) |
| Geltinger Birk | 0 | 4 (44.4%) | 2 (22.2%) | 3 (33.3%) |
| Friedrichsgabekoog | 2 (12.5%) | 8 (50%) | 1 (6.3%) | 5 (31.3%) |
| Sylt | 2 (40%) | 3 (60%) | 0 | 0 |
| total | 4 (8.9%) | 23 (51.1%) | 5 (11.1%) | 13 (28.9%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gross, S.; Müller, A.; Seinige, D.; Oliveira, M.; Steinhagen, D.; Siebert, U.; Kehrenberg, C. High Rates of Multidrug-Resistant Escherichia coli in Great Cormorants (Phalacrocorax carbo) of the German Baltic and North Sea Coasts: Indication of Environmental Contamination and a Potential Public Health Risk. Pathogens 2022, 11, 836. https://doi.org/10.3390/pathogens11080836
Gross S, Müller A, Seinige D, Oliveira M, Steinhagen D, Siebert U, Kehrenberg C. High Rates of Multidrug-Resistant Escherichia coli in Great Cormorants (Phalacrocorax carbo) of the German Baltic and North Sea Coasts: Indication of Environmental Contamination and a Potential Public Health Risk. Pathogens. 2022; 11(8):836. https://doi.org/10.3390/pathogens11080836
Chicago/Turabian StyleGross, Stephanie, Anja Müller, Diana Seinige, Manuela Oliveira, Dieter Steinhagen, Ursula Siebert, and Corinna Kehrenberg. 2022. "High Rates of Multidrug-Resistant Escherichia coli in Great Cormorants (Phalacrocorax carbo) of the German Baltic and North Sea Coasts: Indication of Environmental Contamination and a Potential Public Health Risk" Pathogens 11, no. 8: 836. https://doi.org/10.3390/pathogens11080836






