The Effects of Swine Coronaviruses on ER Stress, Autophagy, Apoptosis, and Alterations in Cell Morphology
Abstract
:1. Swine Coronaviruses
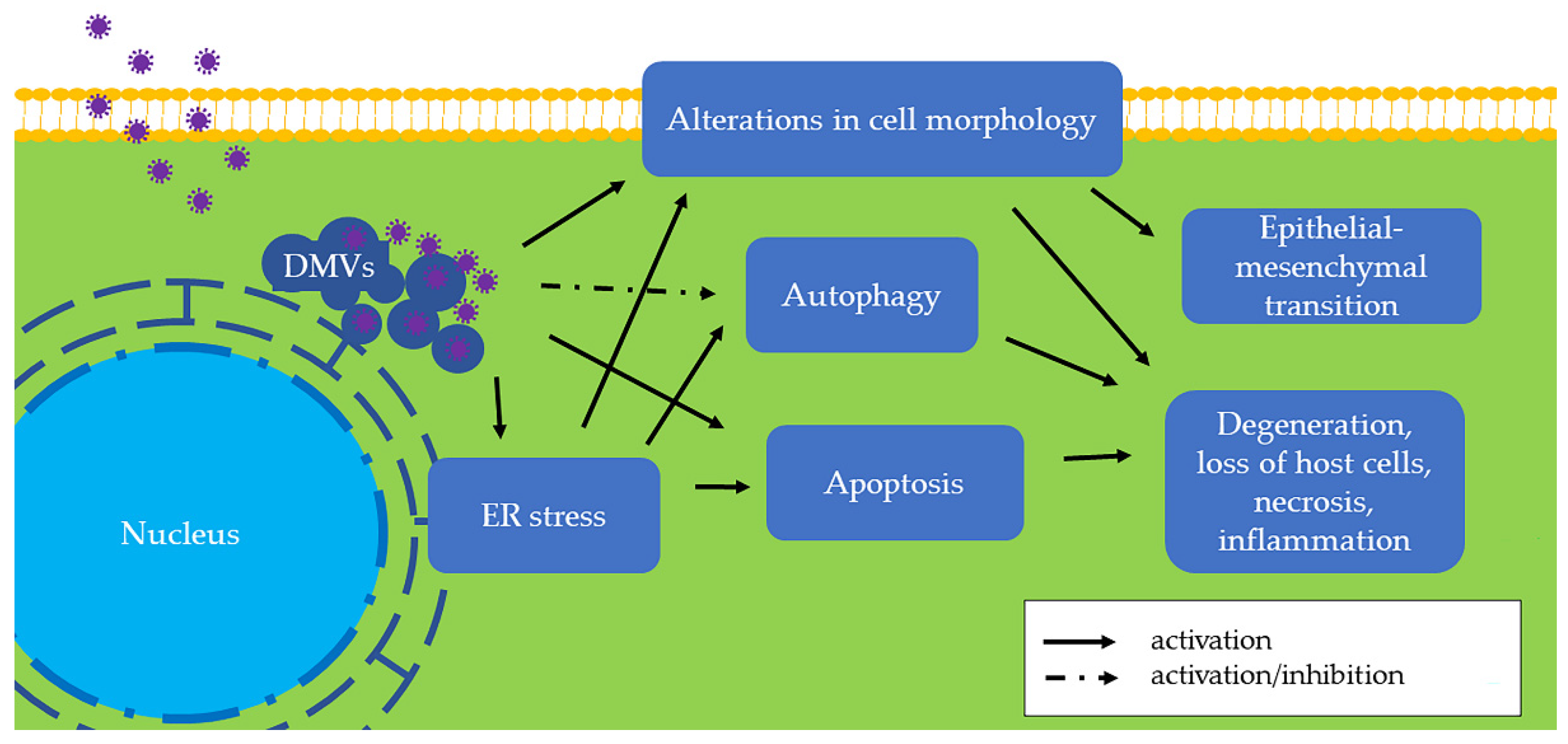
2. ER Stress
3. ER Stress in Swine CoV Infection
| Name of Virus | ER Stress | Autophagy | Apoptosis | Alterations in Cell Morphology |
|---|---|---|---|---|
| PEDV | In vivo Jejunal epithelial cells in 8-week-old pigs [25] In vitro Vero cells [17,26,27] IECs [29,30,31] HEK293T cells [34] | In vitro Vero cells [27,34,50] IPEC-J2 cells [51,52] ST cells [53] HEK293T cells [54] | In vivo Small intestine in 4-day-old piglets [55] Jejunal epithelial cells in 5- and 8-week-old pigs [25,56] In vitro Vero cells [55,57,58,59,60] IECs [61] IPEC-J2 cells [62,63] | In vivo Reduced ZO-1 in jejunum in 4-week-old pigs [64] EMT in jejunum in 4-week-old pigs [64] In vitro Reduced ZO-1 in IPEC-J2 cells [65] Disrupted protein level of AJs and TJs in Vero cells [59] |
| TGEV | In vitro Porcine intestinal epithelial cells [35] ST cells [35] | In vitro IPEC-J2 cells [66] | In vitro IPEC-J2 cells [67] PK-15 cells [68,69,70,71,72] ST cells [73,74,75] | In vitro Reduced E-cadherin, occluding, ZO-1 in IPEC-J2 cells [65] Microfilament and F-actin reorganization in IPEC-J2 cells [65] EMT in IPEC-J2 cells [76] Elevated microfilament and microtubule in ST cells [77] Altered cytoskeleton and vimentin in ST cells [78] |
| PDCoV | In vitro IPI-2I cells [41] LLC-PK1 cells [41] PK-15 cells [39] | In vitro LLC-PK1 cells [79] | In vivo Jejunal and ileum in 7-day-old pig [80] In vitro LLC-PK1 cells [81,82] ST cells [81,82,83] | In vivo Reduced ZO-1 in small intestine in 7-day-old pigs [80] |
| SADS-CoV | In vitro IPEC-J2 cells [42] | n/a | In vitro Vero cells [84] IPI-2I cells [84] | n/a |
| PHEV | In vivo Mouse brains [44] In vitro N2a cells [44] | In vitro N2a cells [85,86] | In vitro PK-15 cells [87] | In vivo Actin rearrangement in mouse brain [88] In vitro Actin rearrangement in N2a cells [89,90] |
| PRCV | n/a | n/a | n/a | n/a |
4. Autophagy in Swine CoV Infection
5. Apoptosis in Swine CoV Infection
6. Cellular Morphologic Alterations in Swine CoV Infection
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saif, L.J.; Wang, Q.; Vlasova, A.N.; Jung, K.; Xiao, S. Diseases of Swine, 11th ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; Chapter 31 Coronaviruses; pp. 488–523. [Google Scholar]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; et al. Changes to Virus Taxonomy and the Statutes Ratified by the International Committee on Taxonomy of Viruses (2020). Arch. Virol. 2020, 165, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Hu, H.; Saif, L.J. Porcine Deltacoronavirus Infection: Etiology, Cell Culture for Virus Isolation and Propagation, Molecular Epidemiology and Pathogenesis. Virus Res. 2016, 226, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Yu, J.-Q.; Huang, Y.-W. Swine Enteric Alphacoronavirus (Swine Acute Diarrhea Syndrome Coronavirus): An Update Three Years after Its Discovery. Virus Res. 2020, 285, 198024. [Google Scholar] [CrossRef] [PubMed]
- Debouck, P.; Pensaert, M.; Coussement, W. The Pathogenesis of an Enteric Infection in Pigs, Experimentally Induced by the Coronavirus-like Agent, CV 777. Vet. Microbiol. 1981, 6, 157–165. [Google Scholar] [CrossRef]
- Wagner, J.E.; Beamer, P.D.; Ristic, M. Electron Microscopy of Intestinal Epithelial Cells of Piglets Infected with a Transmissible Gastroenteritis Virus. Can. J. Comp. Med. Rev. Can. De Med. Comp. 1973, 37, 177–188. [Google Scholar]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.-L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal Swine Acute Diarrhoea Syndrome Caused by an HKU2-Related Coronavirus of Bat Origin. Nature 2018, 556, 255–259. [Google Scholar] [CrossRef]
- Pan, Y.; Tian, X.; Qin, P.; Wang, B.; Zhao, P.; Yang, Y.-L.; Wang, L.; Wang, D.; Song, Y.; Zhang, X.; et al. Discovery of a Novel Swine Enteric Alphacoronavirus (SeACoV) in Southern China. Vet. Microbiol. 2017, 211, 15–21. [Google Scholar] [CrossRef]
- Oakes, S.A.; Papa, F.R. The Role of Endoplasmic Reticulum Stress in Human Pathology. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 173–194. [Google Scholar] [CrossRef]
- Rao, R.V.; Ellerby, H.M.; Bredesen, D.E. Coupling Endoplasmic Reticulum Stress to the Cell Death Program. Cell Death Differ. 2004, 11, 372–380. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal Integration in the Endoplasmic Reticulum Unfolded Protein Response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Gupta, S.; Samali, A.; Fitzgerald, U.; Deegan, S. Methods for Monitoring Endoplasmic Reticulum Stress and the Unfolded Protein Response. Int. J. Cell Biol. 2010, 2010, 830307. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 MRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Ogata, M.; Hino, S.; Saito, A.; Morikawa, K.; Kondo, S.; Kanemoto, S.; Murakami, T.; Taniguchi, M.; Tanii, I.; Yoshinaga, K.; et al. Autophagy Is Activated for Cell Survival after Endoplasmic Reticulum Stress. Mol. Cell. Biol. 2006, 26, 9220–9231. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Chung, H.-Y.; Bae, C.W.; Kim, C.-J.; Park, S. Ghrelin Suppresses Tunicamycin- or Thapsigargin-Triggered Endoplasmic Reticulum Stress-Mediated Apoptosis in Primary Cultured Rat Cortical Neuronal Cells. Endocr. J. 2011, 58, 409–420. [Google Scholar] [CrossRef]
- Cubillos-Ruiz, J.R.; Bettigole, S.E.; Glimcher, L.H. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell 2017, 168, 692–706. [Google Scholar] [CrossRef]
- Zhou, X.; Cong, Y.; Veenendaal, T.; Klumperman, J.; Shi, D.; Mari, M.; Reggiori, F. Ultrastructural Characterization of Membrane Rearrangements Induced by Porcine Epidemic Diarrhea Virus Infection. Viruses 2017, 9, 251. [Google Scholar] [CrossRef]
- Fung, T.S.; Huang, M.; Liu, D.X. Coronavirus-Induced ER Stress Response and Its Involvement in Regulation of Coronavirus-Host Interactions. Virus Res. 2014, 194, 110–123. [Google Scholar] [CrossRef]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef]
- Boga, J.A.; Coto-Montes, A. ER Stress and Autophagy Induced by SARS-CoV-2: The Targets for Melatonin Treatment. Melatonin Res. 2020, 3, 346–361. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.A.L.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation and Cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef]
- Rashid, F.; Dzakah, E.E.; Wang, H.; Tang, S. The ORF8 Protein of SARS-CoV-2 Induced Endoplasmic Reticulum Stress and Mediated Immune Evasion by Antagonizing Production of Interferon Beta. Virus Res. 2021, 296, 198350. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, J.; Wang, Q.; Liu, X.; Li, X.; Li, P.; Ma, Q.; Cao, C. The Nucleocapsid Protein of Severe Acute Respiratory Syndrome Coronavirus Inhibits Cell Cytokinesis and Proliferation by Interacting with Translation Elongation Factor 1α. J. Virol. 2008, 82, 6962–6971. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Gabler, N.K.; Burrough, E.R. Porcine Epidemic Diarrhea Virus Infection Induces Endoplasmic Reticulum Stress and Unfolded Protein Response in Jejunal Epithelial Cells of Weaned Pigs. Vet. Pathol. 2022, 59, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.R.; Sun, M.X.; Ni, B.; Huan, C.; Huang, L.; Li, C.; Fan, H.J.; Ren, X.F.; Mao, X. Triggering Unfolded Protein Response by 2-Deoxy-d-Glucose Inhibits Porcine Epidemic Diarrhea Virus Propagation. Antivir. Res. 2014, 106, 33–41. [Google Scholar] [CrossRef]
- Sun, P.; Jin, J.; Wang, L.; Wang, J.; Zhou, H.; Zhang, Q.; Xu, X. Porcine Epidemic Diarrhea Virus Infections Induce Autophagy in Vero Cells via ROS-Dependent Endoplasmic Reticulum Stress through PERK and IRE1 Pathways. Vet. Microbiol. 2021, 253, 108959. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Ma, J.; Yu, Z.; Pan, Z.; Lu, C.; Yao, H. Identification of Two Mutation Sites in Spike and Envelope Proteins Mediating Optimal Cellular Infection of Porcine Epidemic Diarrhea Virus from Different Pathways. Vet. Res. 2017, 48, 44. [Google Scholar] [CrossRef]
- Xu, X.G.; Zhang, H.; Zhang, Q.; Dong, J.; Liang, Y.; Huang, Y.; Liu, H.J.; Tong, D.W. Porcine Epidemic Diarrhea Virus E Protein Causes Endoplasmic Reticulum Stress and Up-Regulates Interleukin-8 Expression. Virol. J. 2013, 10, 26. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; Zhang, Q.; Huang, Y.; Dong, J.; Liang, Y.; Liu, H.-J.; Tong, D. Porcine Epidemic Diarrhea Virus N Protein Prolongs S-Phase Cell Cycle, Induces Endoplasmic Reticulum Stress, and up-Regulates Interleukin-8 Expression. Vet. Microbiol. 2013, 164, 212–221. [Google Scholar] [CrossRef]
- Xu, X.G.; Zhang, H.L.; Zhang, Q.; Dong, J.; Huang, Y.; Tong, D.W. Porcine Epidemic Diarrhea Virus M Protein Blocks Cell Cycle Progression at S-Phase and Its Subcellular Localization in the Porcine Intestinal Epithelial Cells. Acta Virol. 2015, 59, 265–275. [Google Scholar] [CrossRef]
- Wei, X.; Li, J.; Zhang, Y.; Gong, L.; Xue, C.; Cao, Y. Profiling of Alternative Polyadenylation and Gene Expression in PEDV-Infected IPEC-J2 Cells. Virus Genes 2021, 57, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, H.; Jin, L.; Wang, L.; Huang, X.; Chen, W.; Yan, M.; Liu, G. Profile Analysis of CircRNAs Induced by Porcine Endemic Diarrhea Virus Infection in Porcine Intestinal Epithelial Cells. Virology 2019, 527, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Xu, J.; Duan, X.; Xu, X.; Li, P.; Cheng, L.; Zheng, L.; Li, X.; Zhang, Y.; Wang, X.; et al. Porcine Epidemic Diarrhea Virus ORF3 Protein Causes Endoplasmic Reticulum Stress to Facilitate Autophagy. Vet. Microbiol. 2019, 235, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Fu, F.; Ma, Y.; Zhang, X.; Li, L.; Feng, L.; Liu, P. The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production. J. Virol. 2018, 92, e00431-18. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, Y.; Chang, R.; Tong, D.; Xu, X. Transmissible Gastroenteritis Virus N Protein Causes Endoplasmic Reticulum Stress, up-Regulates Interleukin-8 Expression and Its Subcellular Localization in the Porcine Intestinal Epithelial Cell. Res. Vet. Sci. 2018, 119, 109–115. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, J.L.; Liang, Y.B.; He, Y.P.; Tong, D.W.; Xu, X.G. Transmissible Gastroenteritis Virus Nsp7 Protein Localized in the Cytoplasm Down-Regulates Interleukin 8 Expression in Porcine Intestinal Epithelial Cell. Acta Virol. 2018, 62, 41–49. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, C.; Xue, M.; Fu, F.; Zhang, X.; Li, L.; Yin, L.; Xu, W.; Feng, L.; Liu, P. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA MiR-30a-5p/SOCS1/3 Axis. J. Virol. 2018, 92, e00728-18. [Google Scholar] [CrossRef]
- Lee, S.; Lee, C. Functional Characterization and Proteomic Analysis of the Nucleocapsid Protein of Porcine Deltacoronavirus. Virus Res. 2015, 208, 136–145. [Google Scholar] [CrossRef]
- Doyle, N.; Hawes, P.C.; Simpson, J.; Adams, L.H.; Maier, H.J. The Porcine Deltacoronavirus Replication Organelle Comprises Double-Membrane Vesicles and Zippered Endoplasmic Reticulum with Double-Membrane Spherules. Viruses 2019, 11, 1030. [Google Scholar] [CrossRef]
- Fang, P.; Tian, L.; Zhang, H.; Xia, S.; Ding, T.; Zhu, X.; Zhang, J.; Ren, J.; Fang, L.; Xiao, S. Induction and Modulation of the Unfolded Protein Response during Porcine Deltacoronavirus Infection. Vet. Microbiol. 2022, 271, 109494. [Google Scholar] [CrossRef]
- Zhang, F.; Yuan, W.; Li, Z.; Zhang, Y.; Ye, Y.; Li, K.; Ding, Z.; Chen, Y.; Cheng, T.; Wu, Q.; et al. RNA-Seq-Based Whole Transcriptome Analysis of IPEC-J2 Cells during Swine Acute Diarrhea Syndrome Coronavirus Infection. Front. Vet. Sci. 2020, 7, 492. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-C.; Bai, W.-Z.; Hirano, N.; Hayashida, T.; Hashikawa, T. Coronavirus Infection of Rat Dorsal Root Ganglia: Ultrastructural Characterization of Viral Replication, Transfer, and the Early Response of Satellite Cells. Virus Res. 2012, 163, 628–635. [Google Scholar] [CrossRef]
- Shi, J.; Li, Z.; Xu, R.; Zhang, J.; Zhou, Q.; Gao, R.; Lu, H.; Lan, Y.; Zhao, K.; He, H.; et al. The PERK/PKR-EIF2α Pathway Negatively Regulates Porcine Hemagglutinating Encephalomyelitis Virus Replication by Attenuating Global Protein Translation and Facilitating Stress Granule Formation. J. Virol. 2021, 96, e0169521. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Guan, J.; He, W.; Lv, X.; Hu, S.; Lan, Y.; Zhao, K.; Gao, F.; Li, F.; Fan, G.; et al. MiR-142a-3p Promotes the Proliferation of Porcine Hemagglutinating Encephalomyelitis Virus by Targeting Rab3a. Arch. Virol. 2020, 165, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.L.G.; Sola, I.; Becares, M.; Alberca, B.; Plana, J.; Enjuanes, L.; Zuñiga, S. Coronavirus Gene 7 Counteracts Host Defenses and Modulates Virus Virulence. PLoS Pathog. 2011, 7, e1002090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sharma, N.R.; Zheng, Z.-M.; Chen, M. Viral Regulation of RNA Granules in Infected Cells. Virol. Sin. 2019, 34, 175–191. [Google Scholar] [CrossRef]
- Pandey, K.; Zhong, S.; Diel, D.G.; Hou, Y.; Wang, Q.; Nelson, E.; Wang, X. GTPase-Activating Protein-Binding Protein 1 (G3BP1) Plays an Antiviral Role against Porcine Epidemic Diarrhea Virus. Vet. Microbiol. 2019, 236, 108392. [Google Scholar] [CrossRef]
- Sun, L.; Chen, H.; Ming, X.; Bo, Z.; Shin, H.-J.; Jung, Y.-S.; Qian, Y. Porcine Epidemic Diarrhea Virus Infection Induces Caspase-8-Mediated G3BP1 Cleavage and Subverts Stress Granules to Promote Viral Replication. J. Virol. 2021, 95, e02344-20. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, M.; Zhang, X.; Tan, X.; Guo, H.; Zeng, W.; Yan, G.; Memon, A.M.; Li, Z.; Zhu, Y.; et al. Porcine Epidemic Diarrhea Virus Induces Autophagy to Benefit Its Replication. Viruses 2017, 9, 53. [Google Scholar] [CrossRef]
- Lin, H.; Li, B.; Liu, M.; Zhou, H.; He, K.; Fan, H. Nonstructural Protein 6 of Porcine Epidemic Diarrhea Virus Induces Autophagy to Promote Viral Replication via the PI3K/Akt/MTOR Axis. Vet. Microbiol. 2020, 244, 108684. [Google Scholar] [CrossRef]
- Wu, Y.-T.; Tan, H.-L.; Huang, Q.; Ong, C.-N.; Shen, H.-M. Activation of the PI3K-Akt-MTOR Signaling Pathway Promotes Necrotic Cell Death via Suppression of Autophagy. Autophagy 2009, 5, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, C.; Zhang, B.; Li, Z.; Zeng, W.; Luo, R.; Cao, J.; Cheng, G.; Fan, S.; He, Q. Differential Expression and Correlation Analysis of MiRNA–MRNA Profiles in Swine Testicular Cells Infected with Porcine Epidemic Diarrhea Virus. Sci. Rep. 2021, 11, 1868. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, K.; Xing, Y.; Tu, J.; Yang, X.; Zhao, Q.; Li, K.; Chen, Z. Coronavirus Membrane-Associated Papain-like Proteases Induce Autophagy through Interacting with Beclin1 to Negatively Regulate Antiviral Innate Immunity. Protein Cell 2014, 5, 912–927. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, C. Porcine Epidemic Diarrhea Virus Induces Caspase-Independent Apoptosis through Activation of Mitochondrial Apoptosis-Inducing Factor. Virology 2014, 460–461, 180–193. [Google Scholar] [CrossRef]
- Curry, S.M.; Schwartz, K.J.; Yoon, K.J.; Gabler, N.K.; Burrough, E.R. Effects of Porcine Epidemic Diarrhea Virus Infection on Nursery Pig Intestinal Function and Barrier Integrity. Vet. Microbiol. 2017, 211, 58–66. [Google Scholar] [CrossRef]
- Yang, L.; Wang, C.; Shu, J.; Feng, H.; He, Y.; Chen, J.; Shu, J. Porcine Epidemic Diarrhea Virus Induces Vero Cell Apoptosis via the P53-PUMA Signaling Pathway. Viruses 2021, 13, 1218. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Su, W.; Wang, J.; Sun, Y.; Zhang, J.; Shang, K.; Chen, Z.; Cheng, S.; Wu, H. Genome-Wide Analysis of Differentially Expressed Genes and the Modulation of PEDV Infection in Vero E6 Cells. Microb. Pathog. 2018, 117, 247–254. [Google Scholar] [CrossRef]
- Guo, X.; Hu, H.; Chen, F.; Li, Z.; Ye, S.; Cheng, S.; Zhang, M.; He, Q. ITRAQ-Based Comparative Proteomic Analysis of Vero Cells Infected with Virulent and CV777 Vaccine Strain-like Strains of Porcine Epidemic Diarrhea Virus. J. Proteom. 2015, 130, 65–75. [Google Scholar] [CrossRef]
- Zhang, X.; Li, P.; Zheng, Q.; Hou, J. Lactobacillus Acidophilus S-Layer Protein-Mediated Inhibition of PEDV-Induced Apoptosis of Vero Cells. Vet. Microbiol. 2019, 229, 159–167. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhao, T.Q.; Xu, D.P.; Zhang, X.; Ji, C.J.; Zhang, D.L. The Influence of Porcine Epidemic Diarrhea Virus on Pig Small Intestine Mucosal Epithelial Cell Function. Arch. Virol. 2019, 164, 83–90. [Google Scholar] [CrossRef]
- Shen, X.; Yin, L.; Pan, X.; Zhao, R.; Zhang, D. Porcine Epidemic Diarrhea Virus Infection Blocks Cell Cycle and Induces Apoptosis in Pig Intestinal Epithelial Cells. Microb. Pathog. 2020, 147, 104378. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, B.; Chen, L.; Ma, Z.; He, K.; Fan, H. Differential Protein Analysis of IPEC-J2 Cells Infected with Porcine Epidemic Diarrhea Virus Pandemic and Classical Strains Elucidates the Pathogenesis of Infection. J. Proteome Res. 2017, 16, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Helm, E.T.; Groeltz-Thrush, J.M.; Gabler, N.K.; Burrough, E.R. Epithelial-Mesenchymal Transition of Absorptive Enterocytes and Depletion of Peyer’s Patch M Cells after PEDV Infection. Virology 2021, 552, 43–51. [Google Scholar] [CrossRef]
- Zhao, S.; Gao, J.; Zhu, L.; Yang, Q. Transmissible Gastroenteritis Virus and Porcine Epidemic Diarrhoea Virus Infection Induces Dramatic Changes in the Tight Junctions and Microfilaments of Polarized IPEC-J2 Cells. Virus Res. 2014, 192, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Mou, C.; Yang, X.; Lin, J.; Yang, Q. Mitophagy in TGEV Infection Counteracts Oxidative Stress and Apoptosis. Oncotarget 2016, 7, 27122–27141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ma, X.; Guo, J.; Mi, M.; Wang, K.; Zhang, C.; Tang, X.; Chang, L.; Huang, Y.; Tong, D. Circular RNA CircEZH2 Suppresses Transmissible Gastroenteritis Coronavirus-Induced Opening of Mitochondrial Permeability Transition Pore via Targeting MiR-22 in IPEC-J2. Int. J. Biol. Sci. 2019, 15, 2051–2064. [Google Scholar] [CrossRef]
- Ding, L.; Zhao, X.; Huang, Y.; Du, Q.; Dong, F.; Zhang, H.; Song, X.; Zhang, W.; Tong, D. Regulation of ROS in Transmissible Gastroenteritis Virus-Activated Apoptotic Signaling. Biochem. Biophys. Res. Commun. 2013, 442, 33–37. [Google Scholar] [CrossRef]
- Ding, L.; Li, J.; Li, W.; Fang, Z.; Li, N.; Wu, S.; Li, J.; Hong, M. P53- and ROS-Mediated AIF Pathway Involved in TGEV-Induced Apoptosis. J. Vet. Med. Sci. 2018, 80, 1775–1781. [Google Scholar] [CrossRef]
- Ding, L.; Xu, X.; Huang, Y.; Li, Z.; Zhang, K.; Chen, G.; Yu, G.; Wang, Z.; Li, W.; Tong, D. Transmissible Gastroenteritis Virus Infection Induces Apoptosis through FasL- and Mitochondria-Mediated Pathways. Vet. Microbiol. 2012, 158, 12–22. [Google Scholar] [CrossRef]
- Huang, Y.; Ding, L.; Li, Z.; Dai, M.; Zhao, X.; Li, W.; Du, Q.; Xu, X.; Tong, D. Transmissible Gastroenteritis Virus Infection Induces Cell Apoptosis via Activation of P53 Signalling. J. Gen. Virol. 2013, 94, 1807–1817. [Google Scholar] [CrossRef]
- Ding, L.; Huang, Y.; Du, Q.; Dong, F.; Zhao, X.; Zhang, W.; Xu, X.; Tong, D. TGEV Nucleocapsid Protein Induces Cell Cycle Arrest and Apoptosis through Activation of P53 Signaling. Biochem. Biophys. Res. Commun. 2014, 445, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Eleouet, J.-F.; Chilmonczyk, S.; Besnardeau, L.; Laude, H. Transmissible Gastroenteritis Coronavirus Induces Programmed Cell Death in Infected Cells through a Caspase-Dependent Pathway. J. Virol. 1998, 72, 4918–4924. [Google Scholar] [CrossRef] [PubMed]
- Eléouët, J.-F.; Slee, E.A.; Saurini, F.; Castagné, N.; Poncet, D.; Garrido, C.; Solary, E.; Martin, S.J. The Viral Nucleocapsid Protein of Transmissible Gastroenteritis Coronavirus (TGEV) Is Cleaved by Caspase-6 and -7 during TGEV-Induced Apoptosis. J. Virol. 2000, 74, 3975–3983. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Bai, X.; Guan, L.; Li, J.; Song, X.; Ma, X.; Guo, J.; Zhang, Z.; Du, Q.; Huang, Y.; et al. MicroRNA-4331 Promotes Transmissible Gastroenteritis Virus (TGEV)-Induced Mitochondrial Damage via Targeting RB1, Upregulating Interleukin-1 Receptor Accessory Protein (IL1RAP), and Activating P38 MAPK Pathway in Vitro. Mol. Cell. Proteom. 2018, 17, 190–204. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Dai, L.; Yu, Q.; Yang, Q. Persistent Transmissible Gastroenteritis Virus Infection Enhances Enterotoxigenic Escherichia Coli K88 Adhesion by Promoting Epithelial-Mesenchymal Transition in Intestinal Epithelial Cells. J. Virol. 2017, 91, e01256-17. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, H.-Y.; Chen, J.-F.; Shi, D.; Lang, H.-W.; Wang, Z.-T.; Feng, L. Identification of Cellular Proteome Using Two-Dimensional Difference Gel Electrophoresis in ST Cells Infected with Transmissible Gastroenteritis Coronavirus. Proteome Sci. 2013, 11, 31. [Google Scholar] [CrossRef]
- An, K.; Fang, L.; Luo, R.; Wang, D.; Xie, L.; Yang, J.; Chen, H.; Xiao, S. Quantitative Proteomic Analysis Reveals That Transmissible Gastroenteritis Virus Activates the JAK-STAT1 Signaling Pathway. J. Proteome Res. 2014, 13, 5376–5390. [Google Scholar] [CrossRef]
- Duan, C.; Wang, J.; Liu, Y.; Zhang, J.; Si, J.; Hao, Z.; Wang, J. Antiviral Effects of Ergosterol Peroxide in a Pig Model of Porcine Deltacoronavirus (PDCoV) Infection Involves Modulation of Apoptosis and Tight Junction in the Small Intestine. Vet. Res. 2021, 52, 86. [Google Scholar] [CrossRef]
- Qin, P.; Du, E.-Z.; Luo, W.-T.; Yang, Y.-L.; Zhang, Y.-Q.; Wang, B.; Huang, Y.-W. Characteristics of the Life Cycle of Porcine Deltacoronavirus (PDCoV) in Vitro: Replication Kinetics, Cellular Ultrastructure and Virion Morphology, and Evidence of Inducing Autophagy. Viruses 2019, 11, 455. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, L.; Zhou, P.; Zhang, Y.; Wei, Y.; Wang, Y.; Liu, X. Tandem Mass Tag-Based Quantitative Proteome Analysis of Porcine Deltacoronavirus (PDCoV)-Infected LLC Porcine Kidney Cells. ACS Omega 2020, 5, 21979–21987. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Porcine Deltacoronavirus Induces Apoptosis in Swine Testicular and LLC Porcine Kidney Cell Lines in Vitro but Not in Infected Intestinal Enterocytes in Vivo. Vet. Microbiol. 2016, 182, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Lee, C. Porcine Deltacoronavirus Induces Caspase-Dependent Apoptosis through Activation of the Cytochrome c -Mediated Intrinsic Mitochondrial Pathway. Virus Res. 2018, 253, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Sirinarumitr, T.; Kluge, J.P.; Paul, P.S. Transmissible Gastroenteritis Virus Induced Apoptosis in Swine Testes Cell Cultures. Arch. Virol. 1998, 143, 2471–2485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, Y.; Shi, H.; Chen, J.; Zhang, X.; Wang, X.; Zhou, L.; Liu, J.; Zhang, J.; Ji, Z.; et al. Swine Acute Diarrhea Syndrome Coronavirus-Induced Apoptosis Is Caspase- and Cyclophilin D- Dependent. Emerg. Microbes Infect. 2020, 9, 439–456. [Google Scholar] [CrossRef]
- Li, Z.; Gao, F.; Lan, Y.; Guan, J.; Zhang, J.; Lu, H.; Zhao, K.; He, W. Porcine Hemagglutinating Encephalomyelitis Virus Triggers Neural Autophagy Independently of ULK1. J. Virol. 2021, 95, JVI0085121. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.; Zhao, K.; Lan, Y.; Li, Z.; Lv, X.; Su, J.; Lu, H.; Gao, F.; He, W. Induction of Atypical Autophagy by Porcine Hemagglutinating Encephalomyelitis Virus Contributes to Viral Replication. Front. Cell. Infect. Microbiol. 2017, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Zhao, K.; Wang, G.; Dong, B.; Zhao, J.; Tang, B.; Lu, H.; Gao, W.; Chang, L.; Jin, Z.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Induces Apoptosis in a Porcine Kidney Cell Line via Caspase-Dependent Pathways. Virus Res. 2013, 176, 262–297. [Google Scholar] [CrossRef]
- Lv, X.; Li, Z.; Guan, J.; Zhang, J.; Xu, B.; He, W.; Lan, Y.; Zhao, K.; Lu, H.; Song, D.; et al. ATN-161 Reduces Virus Proliferation in PHEV-Infected Mice by Inhibiting the Integrin A5β1-FAK Signaling Pathway. Vet. Microbiol. 2019, 233, 147–153. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, K.; Lan, Y.; Lv, X.; Hu, S.; Guan, J.; Lu, H.; Zhang, J.; Shi, J.; Yang, Y.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Enters Neuro-2a Cells via Clathrin-Mediated Endocytosis in a Rab5-, Cholesterol-, and PH-Dependent Manner. J. Virol. 2017, 91, e01083-17. [Google Scholar] [CrossRef]
- Lv, X.; Li, Z.; Guan, J.; Hu, S.; Zhang, J.; Lan, Y.; Zhao, K.; Lu, H.; Song, D.; He, H.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Activation of the Integrin A5β1-FAK-Cofilin Pathway Causes Cytoskeletal Rearrangement to Promote Its Invasion of N2a Cells. J. Virol. 2019, 93, e01736-18. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and Molecular Mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Cottam, E.M.; Maier, H.J.; Manifava, M.; Vaux, L.C.; Chandra-Schoenfelder, P.; Gerner, W.; Britton, P.; Ktistakis, N.T.; Wileman, T. Coronavirus Nsp6 Proteins Generate Autophagosomes from the Endoplasmic Reticulum via an Omegasome Intermediate. Autophagy 2011, 7, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Prentice, E.; Jerome, W.G.; Yoshimori, T.; Mizushima, N.; Denison, M.R. Coronavirus Replication Complex Formation Utilizes Components of Cellular Autophagy. J. Biol. Chem. 2004, 279, 10136–10141. [Google Scholar] [CrossRef] [PubMed]
- Kindrachuk, J.; Ork, B.; Hart, B.J.; Mazur, S.; Holbrook, M.R.; Frieman, M.B.; Traynor, D.; Johnson, R.F.; Dyall, J.; Kuhn, J.H.; et al. Antiviral Potential of ERK/MAPK and PI3K/AKT/MTOR Signaling Modulation for Middle East Respiratory Syndrome Coronavirus Infection as Identified by Temporal Kinome Analysis. Antimicrob. Agents Chemother. 2015, 59, 1088–1099. [Google Scholar] [CrossRef]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Kainz, K.; Hofer, S.J.; Kroemer, G.; Madeo, F. Digesting the Crisis: Autophagy and Coronaviruses. Microb. Cell 2020, 7, 119–128. [Google Scholar] [CrossRef]
- Liang, Q.; Seo, G.J.; Choi, Y.J.; Kwak, M.-J.; Ge, J.; Rodgers, M.A.; Shi, M.; Leslie, B.J.; Hopfner, K.-P.; Ha, T.; et al. Crosstalk between the CGAS DNA Sensor and Beclin-1 Autophagy Protein Shapes Innate Antimicrobial Immune Responses. Cell Host Microbe 2014, 15, 228–238. [Google Scholar] [CrossRef]
- Xing, Y.; Liqi, Z.; Jian, L.; Qinghua, Y.; Qian, Y. Doxycycline Induces Mitophagy and Suppresses Production of Interferon-β in IPEC-J2 Cells. Front. Cell. Infect. Microbiol. 2017, 7, 21. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Zhang, Y.; Yang, H. Quantitative Proteomic Analysis of Porcine Intestinal Epithelial Cells Infected with Porcine Deltacoronavirus Using ITRAQ-Coupled LC-MS/MS. J. Proteome Res. 2020, 19, 4470–4485. [Google Scholar] [CrossRef]
- Wang, Z.; Song, D.; Wang, G.; Li, C.; Liu, X.; Wang, X.; Li, Z.; Guan, J.; Zhao, K.; He, W.; et al. Porcine Hemagglutinating Encephalomyelitis Virus Induces Atypical Autophagy via Opposite Regulation of Expression and Nuclear Translocation of Transcription Factor EB. Vet. Microbiol. 2021, 255, 109015. [Google Scholar] [CrossRef]
- Lan, Y.; Li, Z.; Wang, Z.; Wang, X.; Wang, G.; Zhang, J.; Hu, S.; Zhao, K.; Xu, B.; Gao, F.; et al. An Experimental Model of Neurodegenerative Disease Based on Porcine Hemagglutinating Encephalomyelitis Virus–Related Lysosomal Abnormalities. Mol. Neurobiol. 2020, 57, 5299–5306. [Google Scholar] [CrossRef]
- Ko, S.; Gu, M.J.; Kim, C.G.; Kye, Y.C.; Lim, Y.; Lee, J.E.; Park, B.C.; Chu, H.; Han, S.H.; Yun, C.H. Rapamycin-Induced Autophagy Restricts Porcine Epidemic Diarrhea Virus Infectivity in Porcine Intestinal Epithelial Cells. Antivir. Res. 2017, 146, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, H.; Gu, W.; Luo, X.; Li, R.; Zhang, J.; Xu, Y.; Yang, L.; Shen, N.; Feng, L.; et al. Autophagy Negatively Regulates Transmissible Gastroenteritis Virus Replication. Sci. Rep. 2016, 6, 23864. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Liu, Y.; Hao, Z.; Wang, J. Ergosterol Peroxide Suppresses Porcine Deltacoronavirus (PDCoV)-Induced Autophagy to Inhibit Virus Replication via P38 Signaling Pathway. Vet. Microbiol. 2021, 257, 109068. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Peng, O.; Sun, R.; Xu, Q.; Hu, F.; Zhao, Y.; Xue, C.; Cao, Y.; Zhang, H. Transcriptional Landscape of Vero E6 Cells during Early Swine Acute Diarrhea Syndrome Coronavirus Infection. Viruses 2021, 13, 674. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Martin, S.J. Caspase-Independent Cell Death. Nat. Med. 2005, 11, 725–730. [Google Scholar] [CrossRef]
- Lorenzo, H.K.; Susin, S.A.; Penninger, J.; Kroemer, G. Apoptosis Inducing Factor (AIF): A Phylogenetically Old, Caspase-Independent Effector of Cell Death. Cell Death Differ. 1999, 6, 516–524. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Kim, D.-S.; Kim, J.-H.; Lee, G.-H.; Kim, H.-T.; Lim, J.M.; Chae, S.-W.; Chae, H.-J.; Kim, H.-R. P38 Mitogen-Activated Protein Kinase Is Involved in Endoplasmic Reticulum Stress-Induced Cell Death and Autophagy in Human Gingival Fibroblasts. Biol. Pharm. Bull. 2010, 33, 545–549. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Guan, Z.; Li, H.; Ye, M.; Chen, X.; Shen, J.; Zhou, Y.; Shi, Z.-L.; Zhou, P.; et al. SARS-CoV-2 Triggers Inflammatory Responses and Cell Death through Caspase-8 Activation. Signal Transduct. Target. Ther. 2020, 5, 235. [Google Scholar] [CrossRef]
- Lee, C.; Kim, Y.; Jeon, J.H. JNK and P38 Mitogen-Activated Protein Kinase Pathways Contribute to Porcine Epidemic Diarrhea Virus Infection. Virus Res. 2016, 222, 1–12. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, C. Stress-Activated Protein Kinases Are Involved in the Replication of Porcine Deltacoronavirus. Virology 2021, 559, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, Y.; Zhang, Q.; Yang, F.; Yin, Z.; Wang, L.; Li, Q. Porcine Epidemic Diarrhea Virus Infections Induce Apoptosis in Vero Cells via a Reactive Oxygen Species (ROS)/P53, but Not P38 MAPK and SAPK/JNK Signalling Pathways. Vet. Microbiol. 2019, 232, 1–12. [Google Scholar] [CrossRef]
- Zeng, S.; Zhang, H.; Ding, Z.; Luo, R.; An, K.; Liu, L.; Bi, J.; Chen, H.; Xiao, S.; Fang, L. Proteome Analysis of Porcine Epidemic Diarrhea Virus (PEDV)-Infected Vero Cells. PROTEOMICS 2015, 15, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Kim, Y.; Chang, K.-O. Caspase-Mediated Cleavage of Nucleocapsid Protein of a Protease-Independent Porcine Epidemic Diarrhea Virus Strain. Virus Res. 2020, 285, 198026. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Z.; Li, J.; Gao, Y.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H. Porcine Epidemic Diarrhea Virus S1 Protein Is the Critical Inducer of Apoptosis. Virol. J. 2018, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Hu, X.; Wang, C.; Chen, B.; Wang, R.; Dong, S.; Yu, R.; Li, Z. Porcine Epidemic Diarrhea Virus (PEDV) ORF3 Enhances Viral Proliferation by Inhibiting Apoptosis of Infected Cells. Viruses 2020, 12, 214. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Jin, Y.; Yang, Q. Antiviral Activity of Interleukin-11 as a Response to Porcine Epidemic Diarrhea Virus Infection. Vet. Res. 2019, 50, 111. [Google Scholar] [CrossRef]
- Zhao, X.; Song, X.; Bai, X.; Fei, N.; Huang, Y.; Zhao, Z.; Du, Q.; Zhang, H.; Zhang, L.; Tong, D. MiR-27b Attenuates Apoptosis Induced by Transmissible Gastroenteritis Virus (TGEV) Infection via Targeting Runt-Related Transcription Factor 1 (RUNX1). PeerJ 2016, 4, e1635. [Google Scholar] [CrossRef]
- Calvo, E.; Escors, D.; López, J.A.; González, J.M.; Álvarez, A.; Arza, E.; Enjuanes, L. Phosphorylation and Subcellular Localization of Transmissible Gastroenteritis Virus Nucleocapsid Protein in Infected Cells. J. Gen. Virol. 2005, 86, 2255–2267. [Google Scholar] [CrossRef]
- Zhao, X.; Song, X.; Bai, X.; Tan, Z.; Ma, X.; Guo, J.; Zhang, Z.; Du, Q.; Huang, Y.; Tong, D. MicroRNA-222 Attenuates Mitochondrial Dysfunction during Transmissible Gastroenteritis Virus Infection. Mol. Cell. Proteom. 2019, 18, 51–64. [Google Scholar] [CrossRef]
- Ding, L.; Li, J.; Li, W.; Fang, Z.; Li, N.; Guo, Q.; Qu, H.; Feng, D.; Li, J.; Hong, M. P53 Mediated IFN-β Signaling to Affect Viral Replication upon TGEV Infection. Vet. Microbiol. 2018, 227, 61–68. [Google Scholar] [CrossRef]
- Ding, L.; Huang, Y.; Dai, M.; Zhao, X.; Du, Q.; Dong, F.; Wang, L.; Huo, R.; Zhang, W.; Xu, X.; et al. Transmissible Gastroenteritis Virus Infection Induces Cell Cycle Arrest at S and G2/M Phases via P53-Dependent Pathway. Virus Res. 2013, 178, 241–251. [Google Scholar] [CrossRef]
- Kim, B.; Kim, O.; Tai, J.H.; Chae, C. Transmissible Gastroenteritis Virus Induces Apoptosis in Swine Testicular Cell Lines but Not in Intestinal Enterocytes. J. Comp. Pathol. 2000, 123, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Luo, S.; Wu, W.; Hu, J.; Zhou, R. Activation of Interleukin-1β Release and Pyroptosis by Transmissible Gastroenteritis Virus Is Dependent on the NOD-like Receptor Protein 3 Inflammasome in Porcine Intestinal Epithelial Cell Line. Viral Immunol. 2021, 34, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Miyazaki, A.; Hu, H.; Saif, L.J. Susceptibility of Porcine IPEC-J2 Intestinal Epithelial Cells to Infection with Porcine Deltacoronavirus (PDCoV) and Serum Cytokine Responses of Gnotobiotic Pigs to Acute Infection with IPEC-J2 Cell Culture-Passaged PDCoV. Vet. Microbiol. 2018, 221, 49–58. [Google Scholar] [CrossRef]
- Cruz-Pulido, D.; Boley, P.A.; Ouma, W.Z.; Alhamo, M.A.; Saif, L.J.; Kenney, S.P. Comparative Transcriptome Profiling of Human and Pig Intestinal Epithelial Cells after Porcine Deltacoronavirus Infection. Viruses 2021, 13, 292. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms Regulating Intestinal Barrier Integrity and Its Pathological Implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.Y.; Alden, S.L.; Funderburg, N.T.; Fu, P.; Levine, A.D. Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals. PLoS Pathog. 2014, 10, e1004198. [Google Scholar] [CrossRef]
- Greber, U.F.; Way, M. A Superhighway to Virus Infection. Cell 2006, 124, 741–754. [Google Scholar] [CrossRef]
- Cortese, M.; Lee, J.-Y.; Cerikan, B.; Neufeldt, C.J.; Oorschot, V.M.J.; Köhrer, S.; Hennies, J.; Schieber, N.L.; Ronchi, P.; Mizzon, G.; et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe 2020, 28, 853–866. [Google Scholar] [CrossRef]
- Adil, M.S.; Khulood, D.; Narayanan, S.P.; Somanath, P.R. Bioinformatics Analyses Reveal Cell-Barrier Junction Modulations in Lung Epithelial Cells on SARS-CoV-2 Infection. Tissue Barriers 2021, 10, 2000300. [Google Scholar] [CrossRef] [PubMed]
- Zong, Q.F.; Huang, Y.J.; Wu, L.S.; Wu, Z.C.; Wu, S.L.; Bao, W.B. Effects of Porcine Epidemic Diarrhea Virus Infection on Tight Junction Protein Gene Expression and Morphology of the Intestinal Mucosa in Pigs. Pol. J. Vet. Sci. 2019, 22, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Kang, W.; Li, Y.; Shan, Y.; Wang, S.; Liu, F. Dynamic Dissection of Dynein and Kinesin-1 Cooperatively Mediated Intercellular Transport of Porcine Epidemic Diarrhea Coronavirus along Microtubule Using Single Virus Tracking. Virulence 2021, 12, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Li, Y.; Kang, W.; Wang, X.; Wu, X.; Wang, S.; Liu, F. Real-Time Analysis of Quantum Dot Labeled Single Porcine Epidemic Diarrhea Virus Moving along the Microtubules Using Single Particle Tracking. Sci. Rep. 2019, 9, 1307. [Google Scholar] [CrossRef]
- Hu, W.; Zhu, L.; Yang, X.; Lin, J.; Yang, Q. The Epidermal Growth Factor Receptor Regulates Cofilin Activity and Promotes Transmissible Gastroenteritis Virus Entry into Intestinal Epithelial Cells. Oncotarget 2016, 7, 12206–12221. [Google Scholar] [CrossRef]
- Hara, Y.; Hasebe, R.; Sunden, Y.; Ochiai, K.; Honda, E.; Sakoda, Y.; Umemura, T. Propagation of Swine Hemagglutinating Encephalomyelitis Virus and Pseudorabies Virus in Dorsal Root Ganglia Cells. J. Vet. Med. Sci. 2009, 71, 595–601. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Zhang, H.J.; Zhang, Y.N.; Zhou, H.; Guan, L.; Li, Y.; Sun, M.J. IL-17A Promotes Initiation and Development of Intestinal Fibrosis through EMT. Dig. Dis. Sci. 2018, 63, 2898–2909. [Google Scholar] [CrossRef]
- Nisticò, P.; Bissell, M.J.; Radisky, D.C. Epithelial-Mesenchymal Transition: General Principles and Pathological Relevance with Special Emphasis on the Role of Matrix Metalloproteinases. Cold Spring Harb. Perspect. Biol. 2012, 4, 1–11. [Google Scholar] [CrossRef]
- Gassler, N.; Rohr, C.; Schneider, A.; Kartenbeck, J.; Bach, A.; Obermüller, N.; Otto, H.F.; Autschbach, F. Inflammatory Bowel Disease Is Associated with Changes of Enterocytic Junctions. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, 216–228. [Google Scholar] [CrossRef]
- Tahoun, A.; Mahajan, S.; Paxton, E.; Malterer, G.; Donaldson, D.S.; Wang, D.; Tan, A.; Gillespie, T.L.; O’Shea, M.; Roe, A.J.; et al. Salmonella Transforms Follicle-Associated Epithelial Cells into M Cells to Promote Intestinal Invasion. Cell Host Microbe 2012, 12, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, L.; Bozzini, S.; Frangipane, V.; Percivalle, E.; de Luigi, A.; Violatto, M.B.; Lopez, G.; Gabanti, E.; Carsana, L.; D’Amato, M.; et al. Neutrophil Extracellular Traps Induce the Epithelial-Mesenchymal Transition: Implications in Post-COVID-19 Fibrosis. Front. Immunol. 2021, 12, 663303. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, C.; Zhang, N.; Liu, G. Porcine Endemic Diarrhea Virus Infection Regulates Long Noncoding RNA Expression. Virology 2019, 527, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Gao, Z.; Cao, R.; Yang, K.; Cui, Y.; Li, S.; Meng, X.; He, Q.; Li, Z. Transmissible Gastroenteritis Virus Infection Up-Regulates FcRn Expression via Nucleocapsid Protein and Secretion of TGF-β in Porcine Intestinal Epithelial Cells. Front. Microbiol. 2020, 10, 3085. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, H.; Chen, J.; Shi, D.; Dong, H.; Feng, L. Identification of the Interaction between Vimentin and Nucleocapsid Protein of Transmissible Gastroenteritis Virus. Virus Res. 2015, 200, 56–63. [Google Scholar] [CrossRef]
- Choi, S.; Lee, C. Functional Characterization and Proteomic Analysis of Porcine Deltacoronavirus Accessory Protein NS7. J. Microbiol. Biotechnol. 2019, 29, 1817–1829. [Google Scholar] [CrossRef]
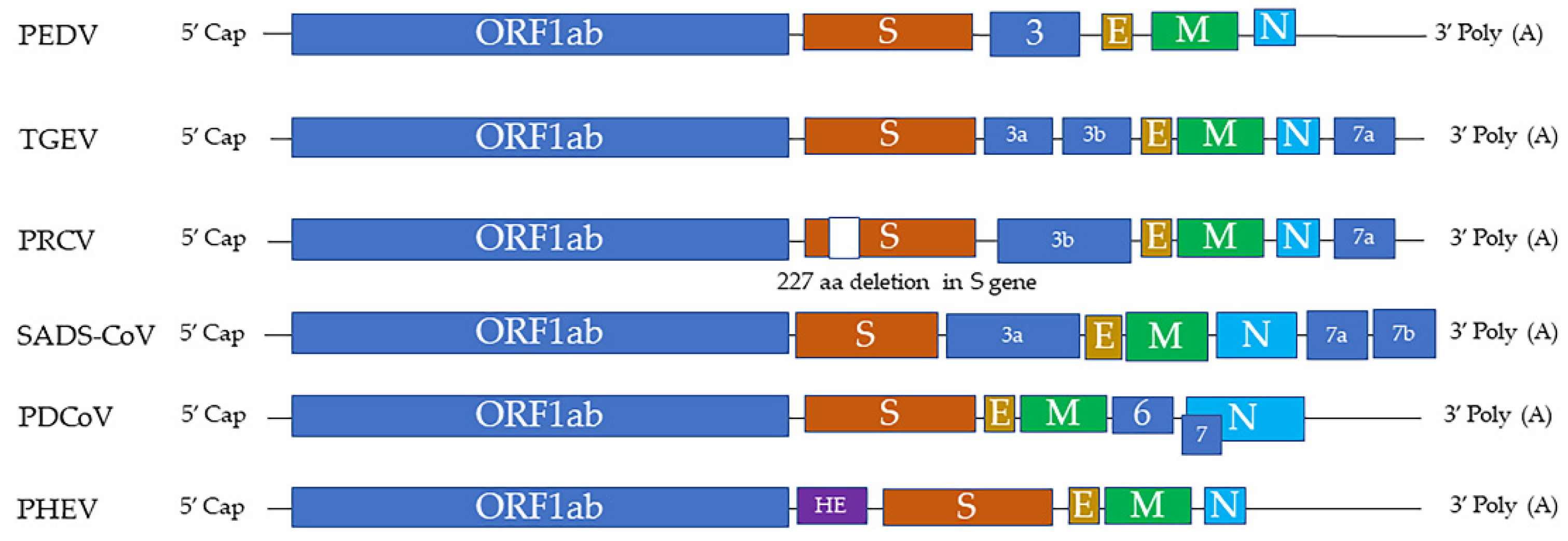
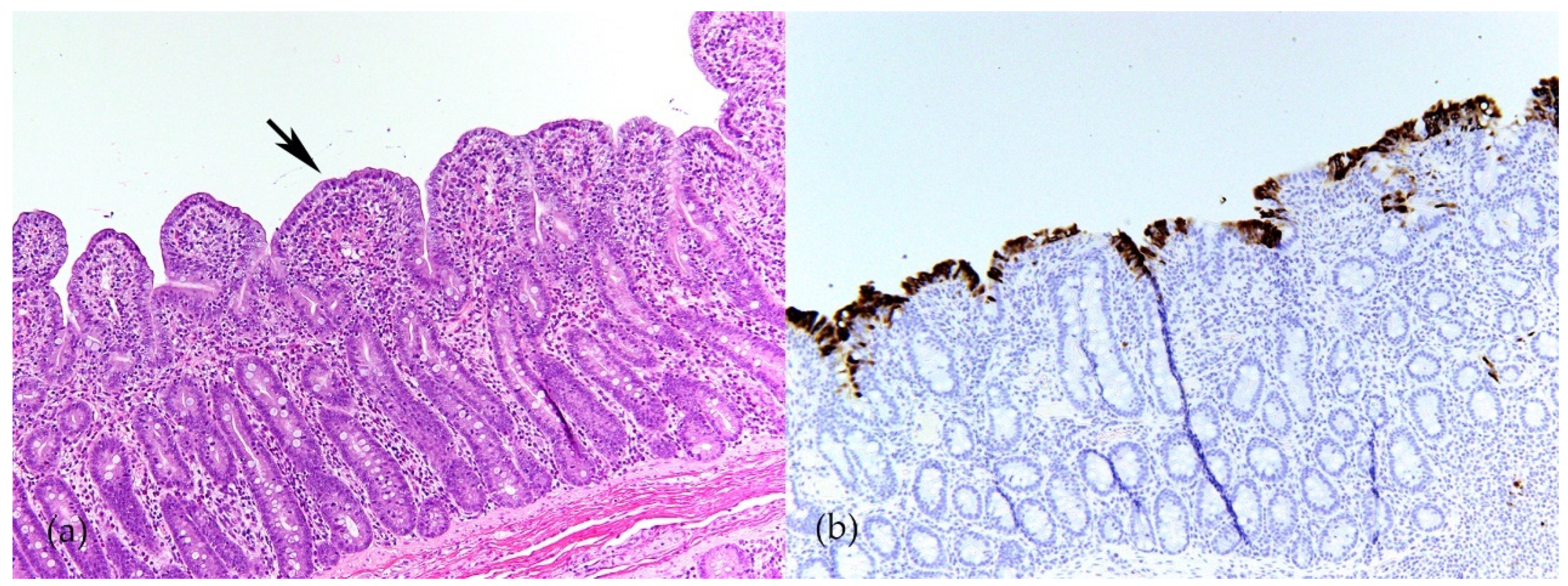
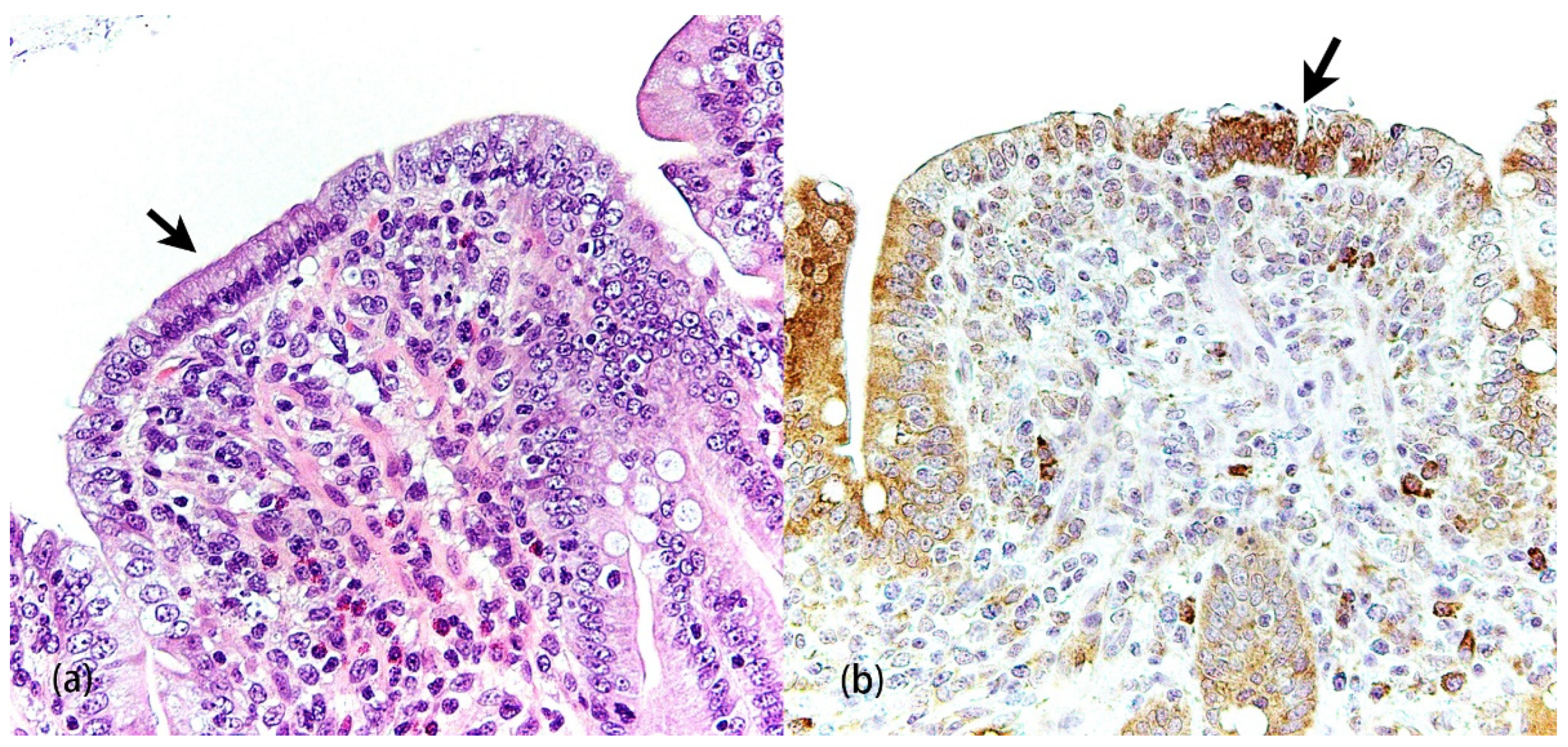
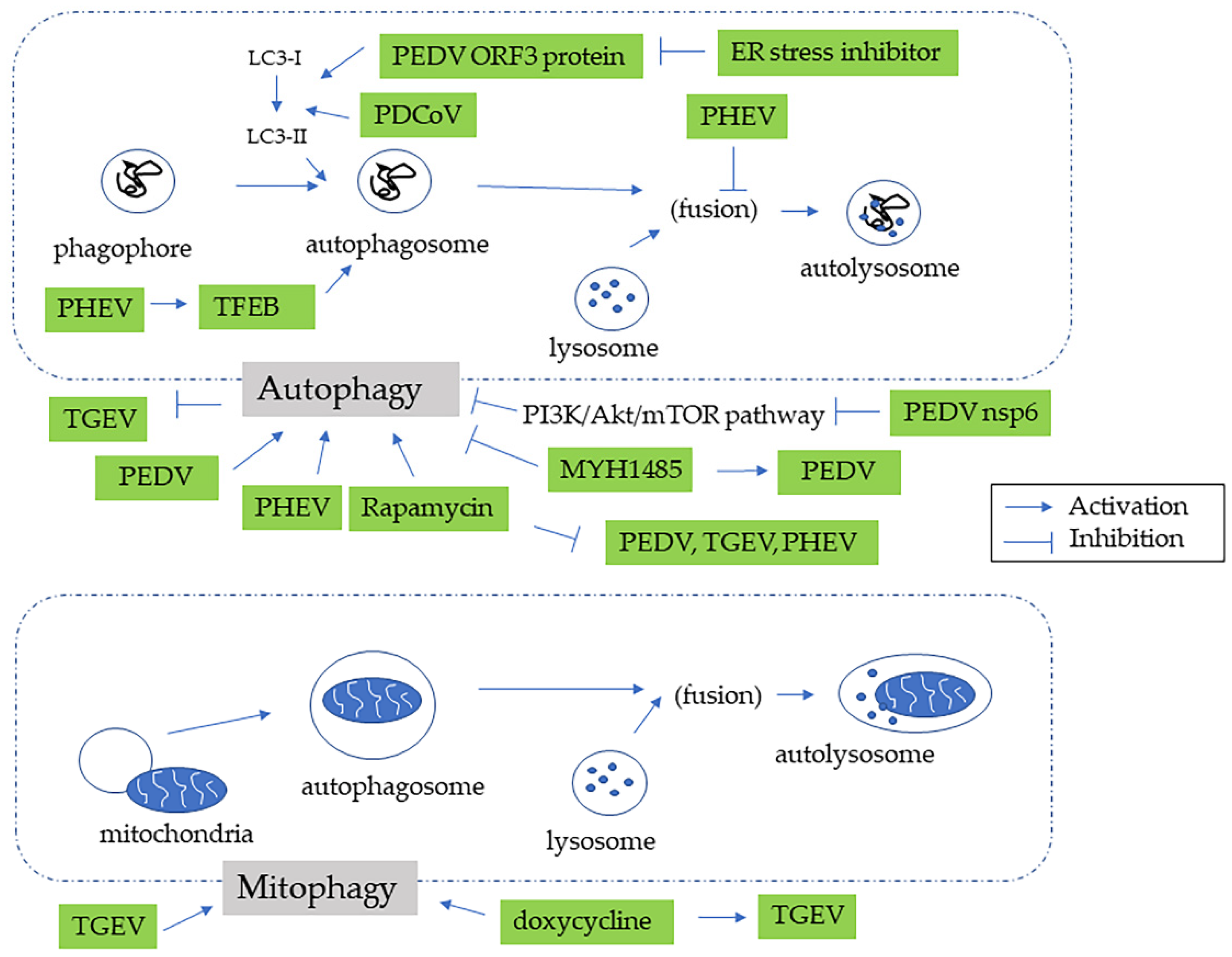
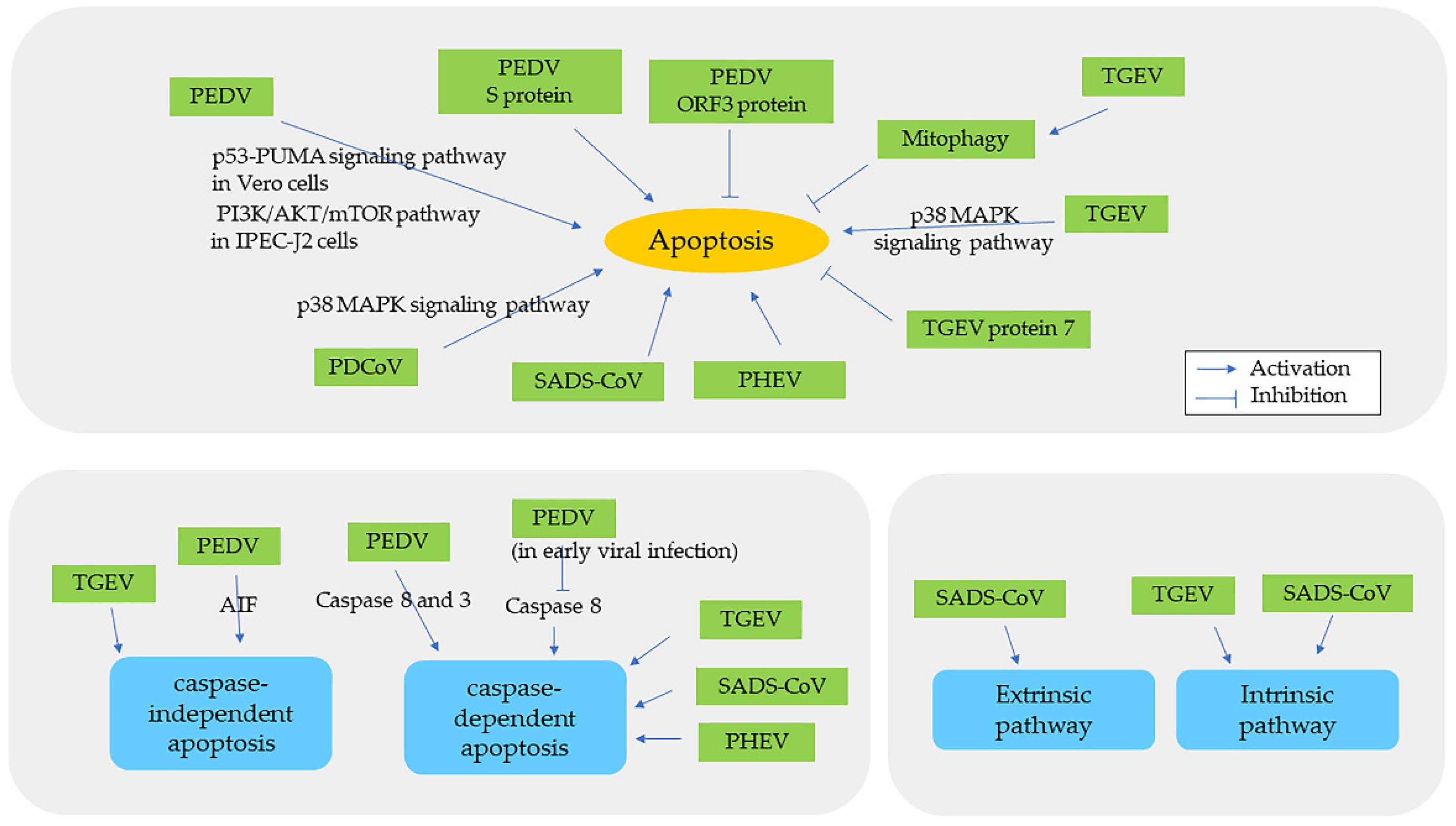
| Name of Virus/ Genera | Major System Affected | Summary and Noteworthy Clinical and Pathological Findings | Ref. |
|---|---|---|---|
| PEDV/ Alphacoronavirus | Enteric | Morbidity: 100% in piglets, less as pigs age Mortality: 50–100% in piglets ≤ 1 week of age Clinical signs: vomiting, watery diarrhea, anorexia, depression Gross lesions: distended small intestine containing yellow fluid and undigested milk Microscopic lesions: villous atrophy in the jejunum and ileum, necrosis of absorptive enterocytes in jejunum | [1] |
| TGEV/ Alphacoronavirus | Enteric | Morbidity: high morbidity in piglets ≤ 2 weeks of age, less as pigs age Mortality: up to 100% in piglets ≤ 2 weeks of age Clinical signs: similar to PEDV Gross lesions: similar to PEDV Microscopic lesions: similar to PEDV | [1] |
| PDCoV/ Deltacoronavirus | Enteric | Morbidity: up to 100% in piglets, less with age Mortality: up to 40% in suckling piglets Clinical signs: similar to PEDV Gross lesions: similar to PEDV and TGEV but less extensive Microscopic lesions: similar to PEDV and TGEV but less extensive | [1] |
| SADS-CoV/ Alphacoronavirus | Enteric | Morbidity: Up to 90% in piglets ≤ 5 days of age, less with age Mortality: over 35% in piglets ≤ 10 days of age Clinical signs: similar to PEDV Gross lesions: similar to PEDV and TGEV but less extensive Microscopic lesions: similar to PEDV and TGEV but less extensive | [7,8] |
| PHEV/ Betacoronavirus | Nervous | Morbidity: up to 100% in neonatal pigs Mortality: up to 100% in neonatal pigs Clinical signs: sneezing or coughing, nervous disorders, vomiting, wasting Gross lesions: cachexia, stomach dilatation, abdominal distension Microscopic lesions: nonsuppurative encephalomyelitis: perivascular cuffing, gliosis, and neuronal degeneration; most pronounced in the gray matter of the pons Varolii, medulla oblongata, and the dorsal horns of the upper spinal cord; degeneration of the ganglia of the stomach wall and perivascular cuffing | [1] |
| PRCV/ Alphacoronavirus | Respiratory | A variant of TGEV with a 227 aa deletion in S gene Morbidity: all ages of pigs can be infected Mortality: minimal (usually subclinical) Clinical signs: coughing, abdominal breathing, dyspnea Gross lesions: mild multifocal consolidation of the lung Microscopic lesions: bronchointerstitial pneumonia, airway epithelial necrosis, type 2 pneumocyte hypertrophy and hyperplasia | [1] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-M.; Burrough, E. The Effects of Swine Coronaviruses on ER Stress, Autophagy, Apoptosis, and Alterations in Cell Morphology. Pathogens 2022, 11, 940. https://doi.org/10.3390/pathogens11080940
Chen Y-M, Burrough E. The Effects of Swine Coronaviruses on ER Stress, Autophagy, Apoptosis, and Alterations in Cell Morphology. Pathogens. 2022; 11(8):940. https://doi.org/10.3390/pathogens11080940
Chicago/Turabian StyleChen, Ya-Mei, and Eric Burrough. 2022. "The Effects of Swine Coronaviruses on ER Stress, Autophagy, Apoptosis, and Alterations in Cell Morphology" Pathogens 11, no. 8: 940. https://doi.org/10.3390/pathogens11080940






