Development of an Orthotopic HPV16-Dependent Base of Tongue Tumor Model in MHC-Humanized Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Mice
2.3. Tumor Implantation
2.4. Western Blot
2.5. siRNA Transfection
2.6. Clonogenic Assay
2.7. Senescence Induction Assay
2.8. Flow Cytometry Assay
2.9. Immunoprecipitation of HLA-Displayed Peptides
2.10. LC-MS
2.11. Cytotoxicity Assay
2.12. Luminescence Detection
2.13. Magnetic Resonance Imaging (MRI)
2.14. Histologic Analysis
3. Results
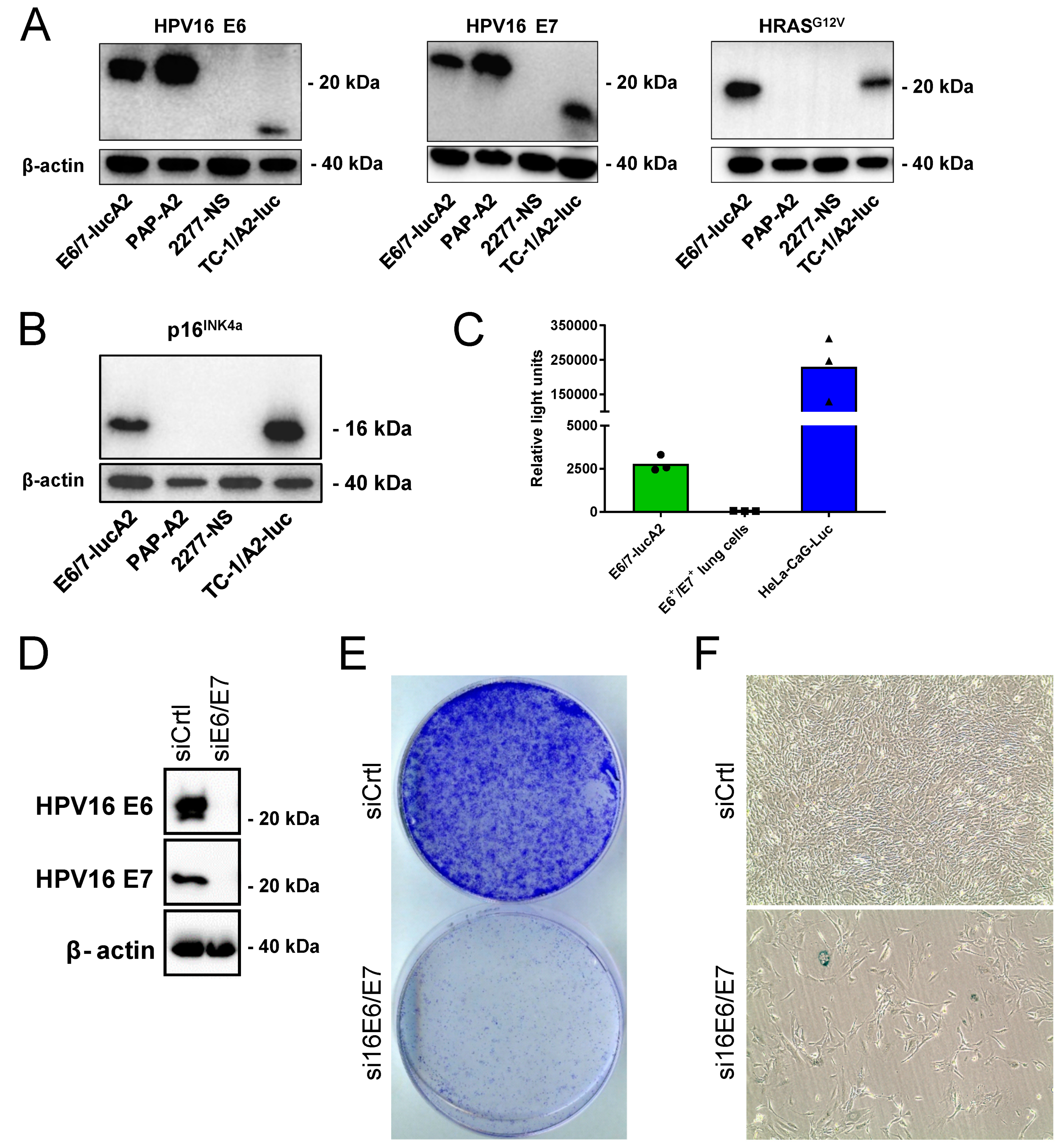
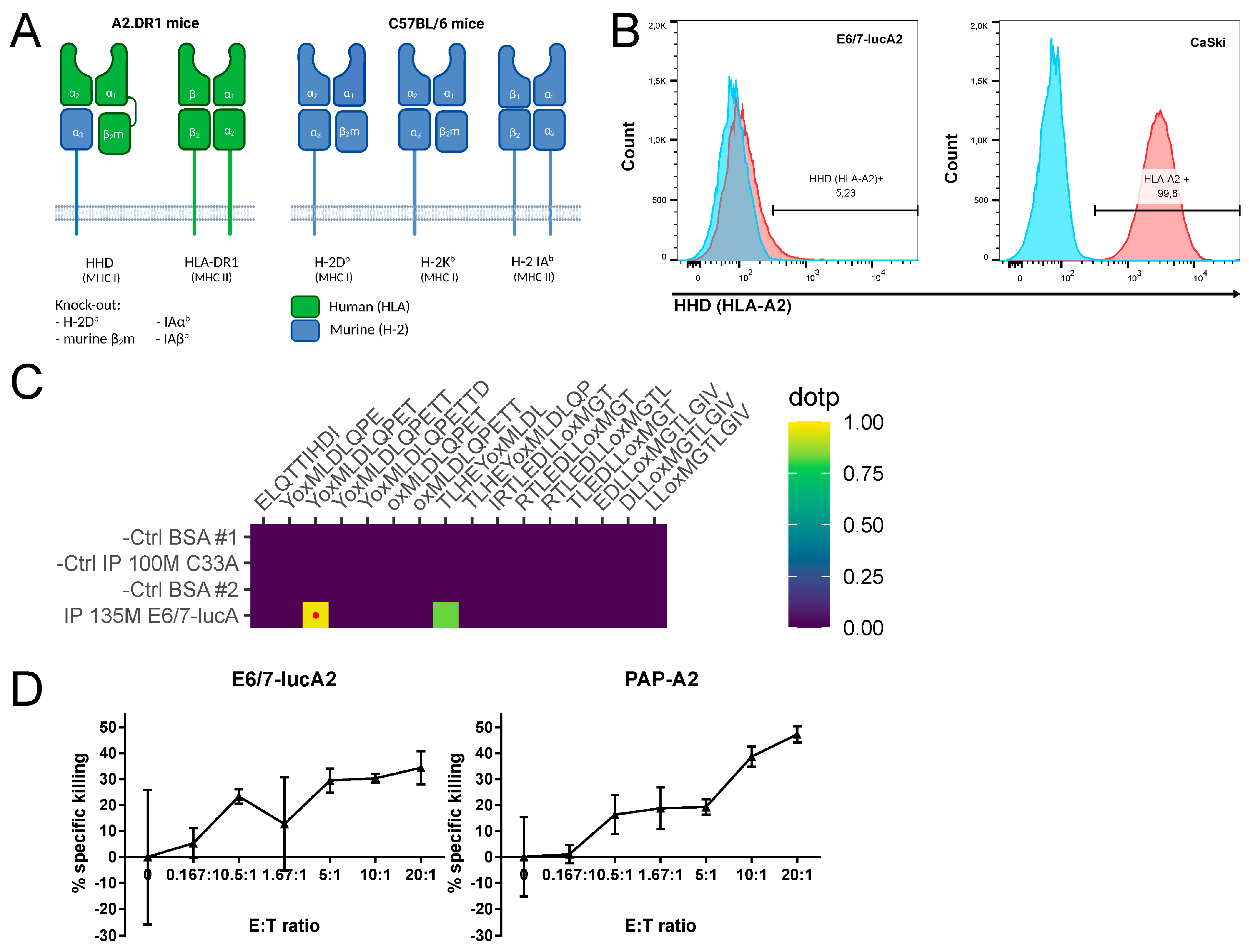
3.1. Generation and Characterization of the E6/7-lucA2 Cell Line
3.2. Analysis of HPV Epitope Presentation by E6/7-lucA2 Cells
3.3. Generation and Characterization of Base of Tongue Tumors Derived from E6/7-lucA2 Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gillison, M.L.; Chaturvedi, A.K.; Anderson, W.F.; Fakhry, C. Epidemiology of Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma. J. Clin. Oncol. 2015, 33, 3235–3242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senkomago, V.; Henley, S.J.; Thomas, C.C.; Mix, J.M.; Markowitz, L.E.; Saraiya, M. Human Papillomavirus-Attributable Cancers—United States, 2012–2016. MMWR Morb. Mortal. Wkly Rep. 2019, 68, 724–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Ozcan-Wahlbrink, M.; Schifflers, C.; Riemer, A.B. Enhanced Radiation Sensitivity of Human Papillomavirus-Driven Head and Neck Cancer: Focus on Immunological Aspects. Front. Immunol. 2019, 10, 2831. [Google Scholar] [CrossRef] [Green Version]
- Tawk, B.; Debus, J.; Abdollahi, A. Evolution of a Paradigm Switch in Diagnosis and Treatment of HPV-Driven Head and Neck Cancer-Striking the Balance Between Toxicity and Cure. Front. Pharmacol. 2021, 12, 753387. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Cheng, M.; Wang, X.; Li, K.; Chen, J.; Chen, Z.; Chen, S.; Chen, J.; Xiong, G.; et al. Tumor microenvironment in head and neck squamous cell carcinoma: Functions and regulatory mechanisms. Cancer Lett. 2021, 507, 55–69. [Google Scholar] [CrossRef]
- Cillo, A.R.; Kurten, C.H.L.; Tabib, T.; Qi, Z.; Onkar, S.; Wang, T.; Liu, A.; Duvvuri, U.; Kim, S.; Soose, R.J.; et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020, 52, 183–199.e9. [Google Scholar] [CrossRef]
- Elmusrati, A.; Wang, J.; Wang, C.Y. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int. J. Oral Sci. 2021, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Tinhofer, I.; Braunholz, D.; Klinghammer, K. Preclinical models of head and neck squamous cell carcinoma for a basic understanding of cancer biology and its translation into efficient therapies. Cancers Head Neck 2020, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Kruse, S.; Buchler, M.; Uhl, P.; Sauter, M.; Scherer, P.; Lan, T.C.T.; Zottnick, S.; Klevenz, A.; Yang, R.; Rösl, F.; et al. Therapeutic vaccination using minimal HPV16 epitopes in a novel MHC-humanized murine HPV tumor model. Oncoimmunology 2019, 8, e1524694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindt, N.; Descamps, G.; Seminerio, I.; Bellier, J.; Lechien, J.R.; Mat, Q.; Pottier, C.; Delvenne, P.; Journé, F.; Saussez, S. High stromal Foxp3-positive T cell number combined to tumor stage improved prognosis in head and neck squamous cell carcinoma. Oral Oncol. 2017, 67, 183–191. [Google Scholar] [CrossRef]
- Manzotti, C.; Audisio, R.A.; Pratesi, G. Importance of orthotopic implantation for human tumors as model systems: Relevance to metastasis and invasion. Clin. Exp. Metastasis 1993, 11, 5–14. [Google Scholar] [CrossRef]
- Ireson, C.R.; Alavijeh, M.S.; Palmer, A.M.; Fowler, E.R.; Jones, H.J. The role of mouse tumour models in the discovery and development of anticancer drugs. Br. J. Cancer 2019, 121, 101–108. [Google Scholar] [CrossRef]
- Zottnick, S.; Voss, A.L.; Riemer, A.B. Inducing Immunity Where It Matters: Orthotopic HPV Tumor Models and Therapeutic Vaccinations. Front. Immunol. 2020, 11, 1750. [Google Scholar] [CrossRef]
- Peng, S.; Trimble, C.; He, L.; Tsai, Y.C.; Lin, C.T.; Boyd, D.A.; Pardoll, D.; Hung, C.-F.; Wu, T.-C. Characterization of HLA-A2-restricted HPV-16 E7-specific CD8(+) T-cell immune responses induced by DNA vaccines in HLA-A2 transgenic mice. Gene Ther. 2006, 13, 67–77. [Google Scholar] [CrossRef]
- Pascolo, S.; Bervas, N.; Ure, J.M.; Smith, A.G.; Lemonnier, F.A.; Perarnau, B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J. Exp. Med. 1997, 185, 2043–2051. [Google Scholar] [CrossRef] [Green Version]
- Pajot, A.; Michel, M.L.; Fazilleau, N.; Pancre, V.; Auriault, C.; Ojcius, D.M.; Lemonnier, F.A.; Lone, Y.-C. A mouse model of human adaptive immune functions: HLA-A2.1-/HLA-DR1-transgenic H-2 class I-/class II-knockout mice. Eur. J. Immunol. 2004, 34, 3060–3069. [Google Scholar] [CrossRef]
- Leitz, J.; Reuschenbach, M.; Lohrey, C.; Honegger, A.; Accardi, R.; Tommasino, M.; Llano, M.; von Knebel Doeberitz, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Oncogenic human papillomaviruses activate the tumor-associated lens epithelial-derived growth factor (LEDGF) gene. PLoS Pathog. 2014, 10, e1003957. [Google Scholar] [CrossRef] [PubMed]
- Honegger, A.; Schilling, D.; Bastian, S.; Sponagel, J.; Kuryshev, V.; Sültmann, H.; Scheffner, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015, 11, e1004712. [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Herrmann, A.L.; Daschle, A.; Kuhn, B.J.; Strobel, T.D.; Lohrey, C.; Bulkescher, J.; Krijgsveld, J.; Hoppe-Seyler, F. Effects of Metformin on the virus/host cell crosstalk in human papillomavirus-positive cancer cells. Int. J. Cancer 2021, 149, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Bassani-Sternberg, M.; Bräunlein, E.; Klar, R.; Engleitner, T.; Sinitcyn, P.; Audehm, S.; Straub, M.; Weber, J.; Slotta-Huspenina, J.; Specht, K.; et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 2016, 7, 13404. [Google Scholar] [CrossRef] [Green Version]
- Chong, C.; Marino, F.; Pak, H.; Racle, J.; Daniel, R.T.; Müller, M.; Gfeller, D.; Coukos, G.; Bassani-Sternberg, M. High-throughput and Sensitive Immunopeptidomics Platform Reveals Profound Interferongamma-Mediated Remodeling of the Human Leukocyte Antigen (HLA) Ligandome. Mol. Cell. Proteom. 2018, 17, 533–548. [Google Scholar] [CrossRef] [Green Version]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Toprak, U.H.; Gillet, L.C.; Maiolica, A.; Navarro, P.; Leitner, A.; Aebersold, R. Conserved Peptide Fragmentation as a Benchmarking Tool for Mass Spectrometers and a Discriminating Feature for Targeted Proteomics. Mol. Cell. Proteom. 2014, 13, 2056–2071. [Google Scholar] [CrossRef] [Green Version]
- RC Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Stanke, J.; Hoffmann, C.; Erben, U.; von Keyserling, H.; Stevanovic, S.; Cichon, G.; Schneider, A.; Kaufmann, A.M. A flow cytometry-based assay to assess minute frequencies of CD8+ T cells by their cytolytic function. J. Immunol. Methods 2010, 360, 56–65. [Google Scholar] [CrossRef]
- Lhotakova, K.; Grzelak, A.; Polakova, I.; Vackova, J.; Smahel, M. Establishment and characterization of a mouse tumor cell line with irreversible downregulation of MHC class I molecules. Oncol. Rep. 2019, 42, 2826–2835. [Google Scholar] [CrossRef]
- Smahel, M.; Sima, P.; Ludvikova, V.; Marinov, I.; Pokorna, D.; Vonka, V. Immunisation with modified HPV16 E7 genes against mouse oncogenic TC-1 cell sublines with downregulated expression of MHC class I molecules. Vaccine 2003, 21, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Guarnieri, F.G.; Staveley-O’Carroll, K.F.; Levitsky, H.I.; August, J.T.; Pardoll, D.M.; Wu, T.-C. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996, 56, 21–26. [Google Scholar] [PubMed]
- Wong, S.P.; Argyros, O.; Coutelle, C.; Harbottle, R.P. Non-viral S/MAR vectors replicate episomally in vivo when provided with a selective advantage. Gene Ther. 2011, 18, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozza, M.; De Roia, A.; Correia, M.P.; Berger, A.; Tuch, A.; Schmidt, A.; Zörnig, I.; Jäger, D.; Schmidt, P.; Harbottle, R.P. A nonviral, nonintegrating DNA nanovector platform for the safe, rapid, and persistent manufacture of recombinant T cells. Sci. Adv. 2021, 7, eabf1333. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; Gwin, T.K.; McLaughlin-Drubin, M.E. p16 in HPV-associated cancers. Oncotarget 2013, 4, 1864–1865. [Google Scholar] [CrossRef]
- Goodwin, E.C.; Yang, E.; Lee, C.J.; Lee, H.W.; DiMaio, D.; Hwang, E.S. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 2000, 97, 10978–10983. [Google Scholar] [CrossRef] [Green Version]
- Hayflick, L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef]
- Riemer, A.B.; Keskin, D.B.; Zhang, G.; Handley, M.; Anderson, K.S.; Brusic, V.; Reinhold, B.; Reinherz, E.L. A conserved E7-derived cytotoxic T lymphocyte epitope expressed on human papillomavirus 16-transformed HLA-A2+ epithelial cancers. J. Biol. Chem. 2010, 285, 29608–29622. [Google Scholar] [CrossRef] [Green Version]
- Blatnik, R.; Mohan, N.; Bonsack, M.; Falkenby, L.G.; Hoppe, S.; Josef, K.; Steinbach, A.; Becker, S.; Nadler, W.M.; Rucevic, M.; et al. A Targeted LC-MS Strategy for Low-Abundant HLA Class-I-Presented Peptide Detection Identifies Novel Human Papillomavirus T-Cell Epitopes. Proteomics 2018, 18, e1700390. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Del Cid-Pavon, G.M.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schifflers, C.; Zottnick, S.; Förster, J.D.; Kruse, S.; Yang, R.; Wiethoff, H.; Bozza, M.; Hoppe-Seyler, K.; Heikenwälder, M.; Harbottle, R.P.; et al. Development of an Orthotopic HPV16-Dependent Base of Tongue Tumor Model in MHC-Humanized Mice. Pathogens 2023, 12, 188. https://doi.org/10.3390/pathogens12020188
Schifflers C, Zottnick S, Förster JD, Kruse S, Yang R, Wiethoff H, Bozza M, Hoppe-Seyler K, Heikenwälder M, Harbottle RP, et al. Development of an Orthotopic HPV16-Dependent Base of Tongue Tumor Model in MHC-Humanized Mice. Pathogens. 2023; 12(2):188. https://doi.org/10.3390/pathogens12020188
Chicago/Turabian StyleSchifflers, Christoph, Samantha Zottnick, Jonas D. Förster, Sebastian Kruse, Ruwen Yang, Hendrik Wiethoff, Matthias Bozza, Karin Hoppe-Seyler, Mathias Heikenwälder, Richard P. Harbottle, and et al. 2023. "Development of an Orthotopic HPV16-Dependent Base of Tongue Tumor Model in MHC-Humanized Mice" Pathogens 12, no. 2: 188. https://doi.org/10.3390/pathogens12020188





