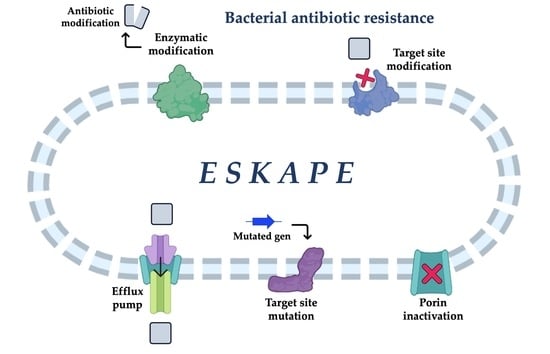
ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic
Abstract
:1. Introduction
2. ESKAPE and Enterobacterales in the COVID Era
3. Acinetobacter baumannii
3.1. Clinical Relevance
3.2. Epidemiology
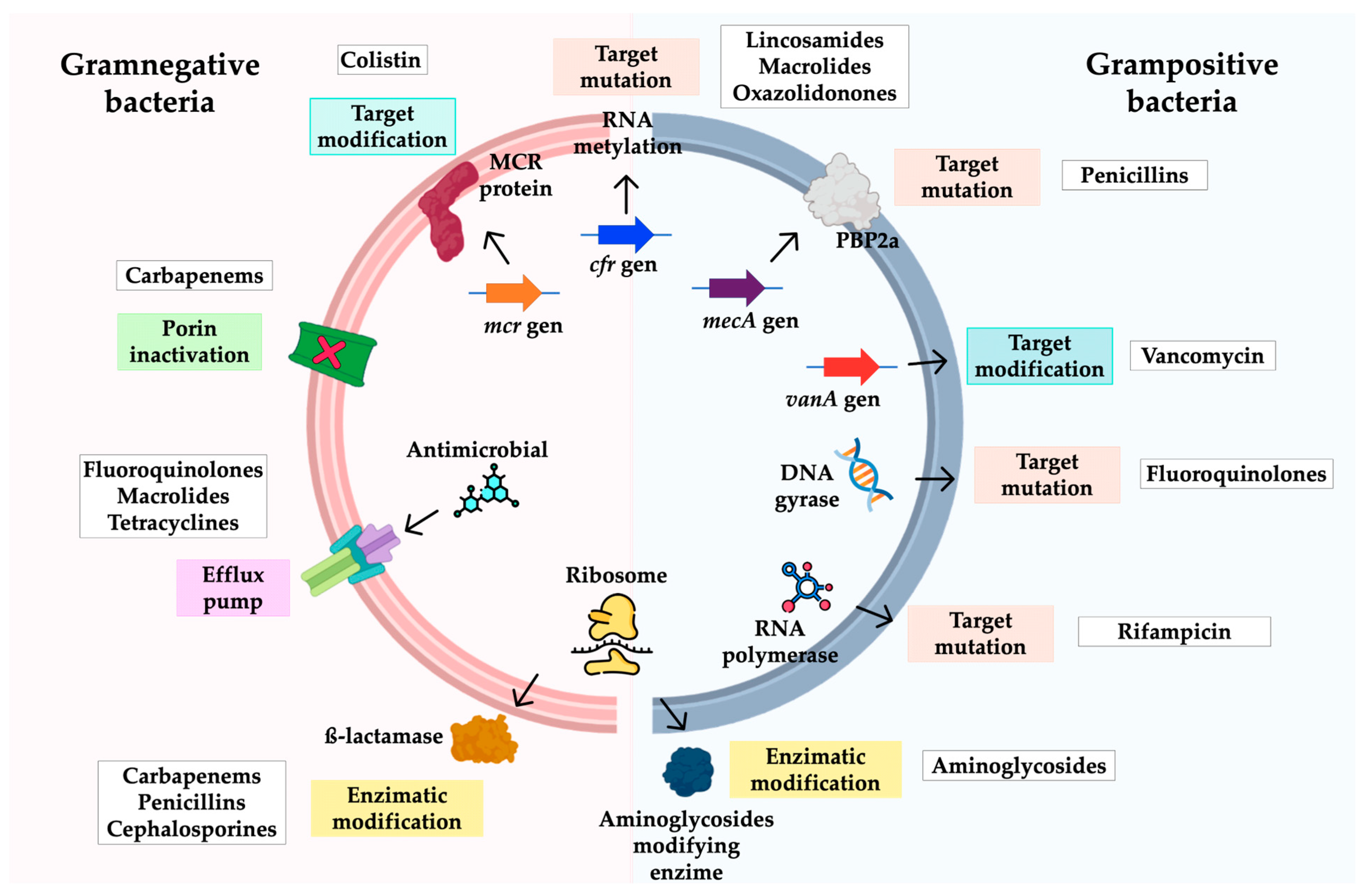
3.3. Mechanisms of Antimicrobial Resistance in A. baumannii
| Resistance Mechanism | Family/Type |
|---|---|
| β-lactamases | TEM (1, 92 *), GES (1, 5, 11, 12, 14), PER (1, 2, 7), CTX-M (2, 5), KPC (2, 10), CARB (4, 10), IMP (1, 2, 4, 5, 6, 8, 11, 19, 24), VIM (1, 2, 3, 4, 11), NDM (1, 2, 3), OXA-2 subgroup (21), OXA-10 subgroup (128), OXA- 20 subgroup (37), OXA-23 subgroup (23), OXA-24 subgroup (133, 239, 24, 25, 26, 40, 72, 143, 182), OXA-51 sub group (51, 64, 65, 66, 68, 70, 71, 69, 75, 76, 77, 79, 80, 104, 106–112, 82, 83, 84, 86, 87, 88, 91, 93, 94, 95, 96, 92, 113), OXA-58 subgroup (58, 96, 97), OXA-143 subgroup (253), OXA-235 subgroup (235) |
| Aminoglycoside-modifying enzymes | Aminoglycoside acetyl-transferases, Aminoglycoside adenylyl-transferases and Aminoglycoside phosphotransferases |
| Permeability defects | AdeABC, AdeFGH, AdeIJK, OmpA, CarO |
3.4. High-Risk Clones
4. Enterobacterales
4.1. Clinical Relevance
4.2. Epidemiology
4.3. Mechanisms of Antibiotic Resistance
4.4. High-Risk Clones
5. Pseudomonas aeruginosa
5.1. Clinical Significance of P. aeruginosa
5.2. Epidemiology
5.3. Resistance Mechanisms
5.4. High-Risk Clones
| Resistance Mechanism | Family/Type |
|---|---|
| β-lactamases | PER (1, 2 *), VEB (2, 3), GES, (2, 5, 18), SHV (2, 5, 12), TEM (4, 21, 24, 42), GES (1, 2, 5, 11, 12, 14, 14, 19, 20, 26, 32), PER (1, 2, 7), CTX-M (1, 2, 3, 14, 15, 43), KPC (2, 5) |
| IMP (1, 2, 4, 5, 6, 7, 9, 10, 11, 13, 14, 15, 16, 18, 19, 20, 21, 22, 25, 26, 29, 30, 31, 33, 35, 37, 40, 41, 43, 44, 45, 48, 56, 62), VIM (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 13, 14, 15, 16, 17, 18, 19, 20, 28, 30, 36, 37, 38), NDM (1, 2) | |
| OXA (OXA-2, OXA-10, OXA-1, OXA-56, OXA-18, OXA-40, OXA-45, OXA-198) | |
| Modification of target site | GyrA/GyrB and ParC/ParE and mcr genes |
| Aminoglycoside-modifying enzymes | Aminoglycoside acetyl-transferases, aminoglycoside adenylyl-transferases and aminoglycoside phosphotransferases |
| Permeability defects | MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM |
| OprD, OprH |
6. Staphylococcus aureus
6.1. Clinical Relevance
6.2. Epidemiology
6.3. Resistance Mechanisms
6.4. High-Risk Clones
7. Enterococcus
7.1. Clinical Relevance
7.2. Epidemiology
7.3. Mechanisms of Antimicrobial Resistance in Enterococcus
7.4. High-Risk Clones
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M. Health care-associated infections- an overview. Infect. Drug. Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Saleem, Z.; Godman, B.; Hassali, M.A.; Hashmi, F.K.; Azhar, F.; Rehman, I.U. Point prevalence surveys of health-care-associated infections: A systematic review. Pathog. Glob. Health 2019, 113, 191–205. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Health Care without Avoidable Infections. The Critical Role of Infection Prevention and Control. 2016. Available online: https://apps.who.int/iris/bitstream/handle/10665/246235/WHO-HIS-SDS-2016.10-eng.pdf (accessed on 16 September 2022).
- Masoudifar, M.; Gouya, M.M.; Pezeshki, Z.; Eshrati, B.; Afhami, S.; Farzami, M.R.; Seifi, A. Health care-associated infections, including device-associated infections, and antimicrobial resistance in Iran: The national update for 2018. J. Prev. Med. Hyg. 2022, 62, E943–E949. [Google Scholar] [CrossRef] [PubMed]
- Blot, S.; Ruppé, E.; Harbarth, S.; Asehnoune, K.; Poulakou, G.; Luyt, C.E.; Rello, J.; Klompas, M.; Depuydt, P.; Eckmann, C.; et al. Healthcare-associated infections in adult intensive care unit patients: Changes in epidemiology, diagnosis, prevention and contributions of new technologies. Intensive Crit. Care Nurs. 2022, 70, 103227. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Burden of AMR Collaborative Group. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Thompson, T. The Staggering Death Toll of Drug-Resistant Bacteria. Nature. Available online: https://www.nature.com/articles/d41586-022-00228-x (accessed on 7 October 2022).
- MacLean, R.C.; Millan, A.S. The evolution of antibiotic resistance. Science 2019, 365, 1082–1083. [Google Scholar] [CrossRef]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017. Available online: https://www.who.int/es/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 7 October 2022).
- Bloom, D.E.; Cadarette, D. Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef]
- Paget, C.; Trottein, F. Mechanisms of Bacterial Superinfection Post-influenza: A Role for Unconventional T Cells. Front. Immunol. 2019, 10, 336. [Google Scholar] [CrossRef]
- Katsurada, N.; Suzuki, M.; Aoshima, M.; Yaegashi, M.; Ishifuji, T.; Asoh, N.; Hamashige, N.; Abe, M.; Ariyoshi, K.; Morimoto, K. The impact of virus infections on pneumonia mortality is complex in adults: A prospective multicentre observational study. BMC Infect. Dis. 2017, 17, 755. [Google Scholar] [CrossRef]
- Johansson, N.; Kalin, M.; Hedlund, J. Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand. J. Infect. Dis. 2011, 43, 609–615. [Google Scholar] [CrossRef]
- Edrada, E.M.; Lopez, E.B.; Villarama, J.B.; Salva Villarama, E.P.; Dagoc, B.F.; Smith, C.; Sayo, A.R.; Verona, J.A.; Trifalgar-Arches, J.; Lazaro, J.; et al. First COVID-19 infections in the Philippines: A case report. Trop. Med. Health 2020, 48, 21. [Google Scholar] [CrossRef] [PubMed]
- Golda, A.; Malek, N.; Dudek, B.; Zeglen, S.; Wojarski, J.; Ochman, M.; Kucewicz, E.; Zembala, M.; Potempa, J.; Pyrc, K. Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells. J. Gen. Virol. 2011, 92, 1358–1368. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, J.A.; Bamford, C.G. SARS-CoV-2, bacterial co-infections, and AMR: The deadly trio in COVID-19? EMBO Mol. Med. 2020, 12, e12560. [Google Scholar] [CrossRef] [PubMed]
- Hendaus, M.A.; Jomha, F.A. COVID-19 induced superimposed bacterial infection. J. Biomol. Struct. Dyn. 2021, 39, 4185–4191. [Google Scholar] [CrossRef]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life 2020, 72, 2097–2111. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Gijón, D.; Ruiz-Garbajosa, P. Antimicrobial resistance in ICUs: An update in the light of the COVID-19 pandemic. Curr. Opin. Crit. Care 2020, 26, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Lansbury, L.; Lim, B.; Baskaran, V.; Lim, W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020, 81, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Kipnis, E.; Sawa, T.; Wiener-Kronish, J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 2006, 36, 78–91. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Imperi, F.; Carattoli, A.; Visca, P. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS. ONE 2011, 6, e22674. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.J.; Loman, N.; Bogaert, D.; O’Grady, J. Co-infections: Potentially lethal and unexplored in COVID-19. Lancet Microbe 2020, 1, e11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Khurana, S.; Singh, P.; Sharad, N.; Kiro, V.V.; Rastogi, N.; Lathwal, A.; Malhotra, R.; Trikha, A.; Mathur, P. Profile of coinfections & secondary infections in COVID-19 patients at a dedicated COVID-19 facility of a tertiary care Indian hospital: Implication on antimicrobial resistance. Ind. J. Med. Microbiol. 2021, 39, 147–153. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Chen, X.; Liao, B.; Cheng, L.; Peng, X.; Xu, X.; Li, Y.; Hu, T.; Li, J.; Zhou, X.; Ren, B. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 7777–7785. [Google Scholar] [CrossRef]
- Rawson, T.M.; Moore, L.; Castro-Sanchez, E.; Charani, E.; Davies, F.; Satta, G.; Ellington, M.J.; Holmes, A.H. COVID-19 and the potential long-term impact on antimicrobial resistance. J. Antimicrob. Chemother. 2020, 75, 1681–1684. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2021, 27, 83–88. [Google Scholar] [CrossRef]
- Rangel, K.; Chagas, T.; De-Simone, S.G. Acinetobacter baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef]
- Cureño-Díaz, M.A.; Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Rojo-Gutiérrez, M.I.; Moncayo-Coello, C.V.; Loyola-Cruz, M.Á.; Castro-Escarpulli, G.; Hernández, D.; Bello-López, J.M. Impact of the modification of a cleaning and disinfection method of mechanical ventilators of COVID-19 patients and ventilator-associated pneumonia: One year experience. Am. J. Infect. Control 2021, 49, 1474–1480. [Google Scholar] [CrossRef]
- Durán-Manuel, E.M.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Bravata-Alcantará, J.C.; Sosa-Hernández, O.; Delgado-Balbuena, L.; León-García, G.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Castro-Escarpulli, G.; et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries. 2021, 15, 58–68. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.; Forde, B.M.; Kidd, T.J.; Harris, P.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Loyola-Cruz, M.Á.; Durán-Manuel, E.M.; Cruz-Cruz, C.; Márquez-Valdelamar, L.M.; Bravata-Alcántara, J.C.; Cortés-Ortíz, I.A.; Cureño-Díaz, M.A.; Ibáñez-Cervantes, G.; Fernández-Sánchez, V.; Castro-Escarpulli, G.; et al. ESKAPE bacteria characterization reveals the presence of Acinetobacter baumannii and Pseudomonas aeruginosa outbreaks in COVID-19/VAP patients. Am. J. Infect. Control 2022, 22, S0196-6553(22)00625-3. [Google Scholar] [CrossRef] [PubMed]
- Touchon, M.; Cury, J.; Yoon, E.J.; Krizova, L.; Cerqueira, G.C.; Murphy, C.; Feldgarden, M.; Wortman, J.; Clermont, D.; Lambert, T. The genomic diversification of the whole Acinetobacter genus: Origins, mechanisms, and consequences. Genome Biol. Evol. 2014, 6, 2866–2882. [Google Scholar] [CrossRef] [PubMed]
- Giammanco, A.; Calà, C.; Fasciana, T.; Dowzicky, M.J. Global Assessment of the Activity of Tigecycline against MultidrugResistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. mSphere 2017, 2, e00310–e00316. [Google Scholar] [CrossRef] [PubMed]
- Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Quirino, A.; Giordano, M.V.; Marano, V.; Rizzo, C.; Liberto, M.C.; Focà, A.; Pavia, M. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect. Dis. 2016, 16, 747. [Google Scholar] [CrossRef]
- Gales, A.C.; Seifert, H.; Gur, D.; Castanheira, M.; Jones, R.N.; Sader, H.S. Antimicrobial Susceptibility of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex and Stenotrophomonas maltophilia Clinical Isolates: Results from the SENTRY Antimicrobial Surveillance Program (1997–2016). Open Forum Infect. Dis. 2019, 6, S34–S46. [Google Scholar] [CrossRef]
- Özgür, E.S.; Horasan, E.S.; Karaca, K.; Ersöz, G.; Naycı Atış, S.; Kaya, A. Ventilator-associated pneumonia due to extensive drug-resistant Acinetobacter baumannii: Risk factors, clinical features, and outcomes. Am. J. Infect. Control 2014, 42, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowski, G.; Pawłowska, I.; Krawczyk, L.; Wojkowska-Mach, J. Antibiotic consumption versus the prevalence of multidrug-resistant Acinetobacter baumannii and Clostridium difficile infections at an ICU from 2014–2015. J. Infect. Public Health 2018, 11, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Mohd Sazlly Lim, S.; Zainal Abidin, A.; Liew, S.M.; Roberts, J.A.; Sime, F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019, 79, 593–600. [Google Scholar] [CrossRef]
- Maes, M.; Higginson, E.; Pereira-Dias, J.; Curran, M.D.; Parmar, S.; Khokhar, F.; Cuchet-Lourenço, D.; Lux, J.; Sharma-Hajela, S.; Ravenhill, B.; et al. Ventilator-associated pneumonia in critically ill patients with COVID-19. Crit. Care 2021, 25, 25. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Yang, Y.; Cai, P.; Cao, J.; Cai, X.; Zhang, Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: A retrospective analysis. Antimicrob. Resist. Infect. Control 2020, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July. Morb. Mortal. Wkly. Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef]
- Ramadan, H.K.-A.; Mahmoud, M.A.; Aburahma, M.Z.; Elkhawaga, A.A.; El-Mokhtar, M.A.; Sayed, I.M.; Hosni, A.; Hassany, S.M.; Medhat, M.A. Predictors of severity and coinfection resistance profile in COVID-19 patients: First report from upper Egypt. Infect. Drug Resist. 2020, 13, 3409–3422. [Google Scholar] [CrossRef]
- Ripa, M.; Galli, L.; Poli, A.; Oltolini, C.; Spagnuolo, V.; Mastrangelo, A.; Muccini, C.; Monti, G.; DeLuca, G.; Landoni, G.; et al. Secondary infections in patients hospitalized with COVID-19: Incidence and predictive factors. Clin. Microbiol. Infect. 2021, 27, 451–457. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Alcántar-Curiel, M.D.; Rosales-Reyes, R.; Jarillo-Quijada, M.D.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Toledano-Tableros, J.E.; Giono-Cerezo, S.; Garza-Villafuerte, P.; López-Huerta, A.; Vences-Vences, D.; et al. Carbapenem-Resistant Acinetobacter baumannii in Three Tertiary Care Hospitals in Mexico: Virulence Profiles, Innate Immune Response and Clonal Dissemination. Front. Microbiol. 2019, 10, 2116. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.T.; Oh, W.S.; Song, J.H.; Chang, H.H.; Jung, S.I.; Kim, S.W.; Ryu, S.Y.; Heo, S.T.; Jung, D.S.; Rhee, J.Y.; et al. Impact of imipenem resistance on mortality in patients with Acinetobacter bacteraemia. J. Antimicrob. Chemother. 2007, 59, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.J.; Domingues, S. Insights on the Horizontal Gene Transfer of Carbapenemase Determinants in the Opportunistic Pathogen Acinetobacter baumannii. Microorganisms 2016, 4, 29. [Google Scholar] [CrossRef]
- David, M.D.; Gill, M.J. Potential for underdosing and emergence of resistance in Acinetobacter baumannii during treatment with colistin. J. Antimicrob. Chemother. 2008, 61, 962–964. [Google Scholar] [CrossRef]
- Gonzalez-Avila, L.U.; Loyola-Cruz, M.A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. Colistin Resistance in Aeromonas spp. Int. J. Mol. Sci. 2021, 22, 5974. [Google Scholar] [CrossRef]
- Martínez-Trejo, A.; Ruiz-Ruiz, J.M.; Gonzalez-Avila, L.U.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Loyola-Cruz, M.A.; Bello-López, J.M.; Castro-Escarpulli, G. Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. Int. J. Mol. Sci. 2022, 23, 6582. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef]
- Rosales-Reyes, R.; Gayosso-Vázquez, C.; Fernández-Vázquez, J.L.; Jarillo-Quijada, M.D.; Rivera-Benítez, C.; Santos-Preciado, J.I.; Alcántar-Curiel, M.D. Virulence profiles and innate immune responses against highly lethal, multidrug-resistant nosocomial isolates of Acinetobacter baumannii from a tertiary care hospital in Mexico. PLoS ONE 2017, 12, e0182899. [Google Scholar] [CrossRef]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Janda, J.M.; Abbott, S.L. The changing face of the family Enterobacteriaceae (order: “Enterobacterales”): New members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 2021, 34, e00174-20. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov. Hafniaceae fam. nov., Morganellaceae fam. nov. fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.J.; Farmer, J.J., III. Enterobacteriaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–24. [Google Scholar] [CrossRef]
- Ordonez, A.A.; Wintaco, L.M.; Mota, F.; Restrepo, A.F.; Ruiz-Bedoya, C.A.; Reyes, C.F.; Jain, S.K. Imaging Enterobacterales infections in patients using pathogen-specific positron emission tomography. Sci. Transl. Med. 2021, 13, eabe9805. [Google Scholar] [CrossRef] [PubMed]
- Sheu, C.C.; Chang, Y.T.; Lin, S.Y.; Chen, Y.H.; Hsueh, P.R. Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front. Microbiol. 2019, 10, 80. [Google Scholar] [CrossRef]
- Yi, J.; Kim, K.H. Identification and infection control of carbapenem-resistant Enterobacterales in intensive care units. Acute Crit. Care 2021, 36, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Gavidia, C.; Álvarez, F.P.; Munita, J.M.; Cortés, S.; Moreno-Switt, A.I. Perspective on Clinically-Relevant Antimicrobial Resistant Enterobacterales in Food: Closing the Gaps Using Genomics. Front. Sustain. Food Syst. 2021, 5, 667504. [Google Scholar] [CrossRef]
- Yu, H.; Hernández González, A.; Estévez Torres, G.; González Molina, M.K.; Hart Casares, M.; Han, X.; Quiñones Pérez, D.A. Retrospective Study of Risk Factors, Mortality, and Treatment Outcomes for Infections with Carbapenemase-Producing Enterobacterales in a Tertiary Hospital in Havana, Cuba. Antibiotics 2022, 11, 942. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Karlowsky, J.A.; de Jonge, B.L.; Stone, G.G.; Sahm, D.F. Epidemiology of carbapenem resistance determinants identified in meropenem-nonsusceptible Enterobacterales collected as part of a global surveillance program, 2012 to 2017. Antimicrob. Agents Chemother. 2021, 65, e02000-20. [Google Scholar] [CrossRef]
- O’Toole, R.F. The interface between COVID-19 and bacterial healthcare-associated infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef]
- Rothe, K.; Feihl, S.; Schneider, J.; Wallnöfer, F.; Wurst, M.; Lukas, M.; Spinner, C.D. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: A retrospective cohort study in light of antibiotic stewardship. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 859–869. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Annavajhala, M.K.; Wang, Z.; Macesic, N.; Hu, Y.; Giddins, M.J.; O’Malley, A.; Toussaint, N.C.; Whittier, S.; Torres, V.J.; et al. Genomic and geographic context for the evolution of high-risk carbapenem-resistant Enterobacter cloacae complex clones ST171 and ST78. mBio 2018, 9, e00542-18. [Google Scholar] [CrossRef]
- Pintos-Pascual, I.; Cantero-Caballero, M.; Rubio, E.M.; Sánchez-Romero, I.; Asensio-Vegas, Á.; Ramos-Martínez, A. Epidemiology and clinic of infections and colonisations caused by carbapenemase-producing Enterobacteriaceae in a tertiary hospital. Rev. Esp. Chemother. 2020, 33, 122. [Google Scholar] [CrossRef]
- Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Occurrence of antibiotic resistance among Enterobacterales isolated from raw and ready-to-eat food-phenotypic and genotypic characteristics. Int. J. Environ. Health Res. 2022, 32, 1733–1744. [Google Scholar] [CrossRef]
- Tumbarello, M.; Losito, A.R.; Giamarellou, H. Optimizing therapy in carbapenem-resistant Enterobacteriaceae infections. Curr. Opin. Infect. Dis. 2018, 31, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.L.; Chen, H.M.; Hii, M.; Hsueh, P.R. Carbapenemase-producing Enterobacterales infections: Recent advances in diagnosis and treatment. Int. J. Antimicrob. Agents 2022, 59, 106528. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Boattini, M.; Comini, S.; Casale, R.; Iannaccone, M.; Cavallo, R.; Costa, C. Occurrence of multi-carbapenemases producers among carbapenemase-producing Enterobacterales and in vitro activity of combinations including cefiderocol, ceftazidime-avibactam, meropenem-vaborbactam, and aztreonam in the COVID-19 era. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 573–580. [Google Scholar] [CrossRef]
- Pintado, V.; Ruiz-Garbajosa, P.; Escudero-Sanchez, R.; Gioia, F.; Herrera, S.; Vizcarra, P.; Fortún, J.; Cobo, J.; Martín-Dávila, P.; Morosini, M.I.; et al. Carbapenemase-producing Enterobacterales infections in COVID-19 patients. Infect. Dis. 2022, 54, 36–45. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965. [Google Scholar] [CrossRef]
- Bardet, L.; Rolain, J.M. Development of new tools to detect colistin-resistance among Enterobacteriaceae strains. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 3095249. [Google Scholar] [CrossRef]
- Partridge, S.R. Resistance mechanisms in Enterobacteriaceae. Pathology 2015, 47, 276–284. [Google Scholar] [CrossRef]
- Gijón, D.; Tedim, A.P.; Valverde, A.; Rodríguez, I.; Morosini, M.I.; Coque, T.M.; Cantón, R. Early OXA-48-producing Enterobacterales isolates recovered in a Spanish hospital reveal a complex introduction dominated by sequence type 11 (ST11) and ST405 Klebsiella pneumoniae clones. mSphere 2020, 5, e00080-20. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Ellaby, N.; Ellington, M.J.; Doumith, M.; Mustafa, N.; Meunier, D.; Woodford, N. Diversity of carbapenemase-producing Enterobacterales in England as revealed by whole-genome sequencing of isolates referred to a national reference laboratory over a 30-month period. J. Med. Microbiol. 2022, 71, 001518. [Google Scholar] [CrossRef]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef]
- Hernández-García, M.; Sánchez-López, J.; Martínez-García, L.; Becerra-Aparicio, F.; Morosini, M.I.; Ruiz-Garbajosa, P.; Cantón, R. Emergence of the new KPC-49 variant conferring an ESBL phenotype with resistance to ceftazidime-avibactam in the ST131-H30R1 Escherichia coli high-risk clone. Pathogens 2021, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, R.; Hussain, A.; Abdullah, A.; Islam, M.; Sadique, M.; Muniruzzaman, S.M.; Tabassum, A.; Halim, F.; Akter, N.; Ahmed, D.; et al. International High-Risk Clones Among Extended-Spectrum β-Lactamase-Producing Escherichia coli in Dhaka, Bangladesh. Front. Microbiol. 2021, 12, 2843. [Google Scholar] [CrossRef]
- Welker, S.; Boutin, S.; Miethke, T.; Heeg, K.; Nurjadi, D. Emergence of carbapenem-resistant ST131 Escherichia coli carrying blaOXA-244 in Germany, 2019 to 2020. Eurosurveillance 2020, 25, 2001815. [Google Scholar] [CrossRef] [PubMed]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.C. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Ben Haj Khalifa, A.; Moissenet, D.; Vu Thien, H.; Khedher, M. Les facteurs de virulence de Pseudomonas aeruginosa: Mécanismes et modes de régulations [Virulence factors in Pseudomonas aeruginosa: Mechanisms and modes of regulation]. Ann. Biol. Clin. 2011, 69, 393–403. [Google Scholar] [CrossRef]
- Qu, J.; Cai, Z.; Liu, Y.; Duan, X.; Han, S.; Liu, J.; Zhu, Y.; Jiang, Z.; Zhang, Y.; Zhuo, C.; et al. Persistent Bacterial Coinfection of a COVID-19 Patient Caused by a Genetically Adapted Pseudomonas aeruginosa Chronic Colonizer. Front. Cell. Infect. Microbiol. 2021, 11, 641920. [Google Scholar] [CrossRef] [PubMed]
- Barrasa, H.; Rello, J.; Tejada, S.; Martín, A.; Balziskueta, G.; Vinuesa, C.; Fernández-Miret, B.; Villagra, A.; Vallejo, A.; San Sebastián, A.; et al. SARS-CoV-2 in Spanish Intensive Care Units: Early experience with 15-day survival in Vitoria. Anaesth. Crit. Care. Pain Med. 2021, 39, 553–561. [Google Scholar] [CrossRef]
- Rhoades, N.S.; Pinski, A.N.; Monsibais, A.N.; Jankeel, A.; Doratt, B.M.; Cinco, I.R.; Ibraim, I.; Messaoudi, I. Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose. Cell Rep. 2021, 36, 109637. [Google Scholar] [CrossRef]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Westblade, L.F.; Simon, M.S.; Satlin, M.J. Bacterial Coinfections in Coronavirus Disease 2019. Trends Microbiol. 2021, 29, 930–941. [Google Scholar] [CrossRef]
- Mahmoudi, H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS Hyg. Infect. Control 2020, 15, Doc35. [Google Scholar] [CrossRef]
- Juan, C.; Torrens, G.; González-Nicolau, M.; Oliver, A. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol. Rev. 2017, 41, 781–815. [Google Scholar] [CrossRef]
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Update 2015, 21–22, 41–59. [Google Scholar] [CrossRef]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef]
- Mulet, X.; Cabot, G.; Ocampo-Sosa, A.A.; Domínguez, M.A.; Zamorano, L.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; Peña, C.; et al. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob. Agents Chemother. 2013, 57, 5527–5535. [Google Scholar] [CrossRef] [PubMed]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Cabot, G.; Rivera, A.; Benito, N.; Segura, C.; Montero, M.M.; Sorlí, L.; Tubau, F.; Gómez-Zorrilla, S.; et al. Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain. Antimicrob. Agents Chemother. 2017, 61, e01589-17. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Garbajosa, P.; Cantón, R. COVID-19: Impact on prescribing and antimicrobial resistance. Rev. Esp. Quimioter. 2021, 34, 63–68. [Google Scholar] [CrossRef]
- López-García, A.; Rocha-Gracia, R.; Bello-López, E.; Juárez-Zelocualtecalt, C.; Sáenz, Y.; Castañeda-Lucio, M.; López-Pliego, L.; González-Vázquez, M.C.; Torres, C.; Ayala-Nuñez, T.; et al. Characterization of antimicrobial resistance mechanisms in carbapenem-resistant Pseudomonas aeruginosa carrying IMP variants recovered from a Mexican Hospital. Infect. Drug Resist. 2018, 11, 1523–1536. [Google Scholar] [CrossRef]
- Lupo, A.; Haenni, M.; Madec, J.Y. Antimicrobial Resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 2018, 6, 3:10. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 100640. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Rasheed, N.A.; Hussein, N.R. Staphylococcus aureus: An Overview of Discovery, Characteristics, Epidemiology, Virulence Factors and Antimicrobial Sensitivity. Eur. J. Mol. Clin. Med. 2021, 8, 1160–1183. [Google Scholar]
- Rachman, A.R.A.; Suhaili, Z.; Desa, M.N.M. The evolution and dissemination of methicillin resistance determinant in Staphylococcus aureus. In The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus; Intech: Vienna, Austria, 2017. [Google Scholar] [CrossRef]
- Sato, A.; Yamaguchi, T.; Hamada, M.; Ono, D.; Sonoda, S.; Oshiro, T.; Nagashima, M.; Kato, K.; Okazumi, S.; Katoh, R.; et al. Morphological and Biological Characteristics of Staphylococcus aureus Biofilm Formed in the Presence of Plasma. Microb. Drug Resist. 2019, 25, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Park, Y.K.; Jung, T.S.; Park, S.B. Molecular Typing, Antibiotic Resistance and Enterotoxin Gene Profiles of Staphylococcus aureus Isolated from Humans in South Korea. Microorganisms 2022, 10, 642. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Jin, H.; Kim, J.; Park, M.; Lee, J.; Kim, S. Molecular Characteristics and Exotoxins of Methicillin-Resistant Staphylococcus aureus. Biomed. Sci. Lett. 2021, 27, 195–207. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M.; Trecarichi, E.M.; Carnelutti, A.; Righi, E.; Del Giacomo, P.; Ansaldi, F.; Trucchi, C.; Alicino, C.; Cauda, R.; et al. Characteristics of Staphylococcus aureus Bacteraemia and Predictors of Early and Late Mortality. PLoS ONE 2017, 12, e0170236. [Google Scholar] [CrossRef]
- The Center for Food Security & Public Health. Available online: https://www.cfsph.iastate.edu (accessed on 7 October 2022).
- Ortíz-Gil, M.Á.; Velazquez-Meza, M.E.; Echániz-Aviles, G.; Mora-Domínguez, J.P.; Carnalla-Barajas, M.N.; Mendiola Del Moral, E.L. Tracking methicillin-resistant Staphylococcus aureus clones in a hospital in Southern Mexico. Salud Publica Mex. 2020, 62, 186–191. [Google Scholar] [CrossRef]
- Cornejo-Juárez, P.; Volkow-Fernández, P.; Sifuentes-Orsonio, J.; Echániz-Aviles, G.; Díaz-González, A.; Velázquez-Acosta, C.; Bobadilla del Valle, M.; Gordillo-Molina, P.; Velazquez-Meza, M.E. Tracing the source of an outbreak of methicillin-resistant Staphylococcus aureus in a tertiary-care oncology hospital by epidemiology and epidemiology. Microb. Drug Resist. 2010, 16, 203–208. [Google Scholar] [CrossRef]
- Espinosa, P.M.; García, F.R.; Mormeneo, B.S.; Martínez, A.R.M.; Frutos, M.V.; Villuendas, U.M.C.; Palacián, R.M.P.; Arbonés, M.J.M.; Martínez, J.C.; Ramos, P.C. Impact of Staphylococcus aureus bacteraemia in patients with COVID-19. Rev. Esp. Chemother. 2022, 35, 468–474. [Google Scholar] [CrossRef]
- Adalbert, J.R.; Varshney, K.; Tobin, R.; Pajaro, R. Clinical outcomes in patients co-infected with COVID-19 and Staphylococcus aureus: A scoping review. BMC Infect. Dis. 2021, 21, 985. [Google Scholar] [CrossRef]
- Chandran, S.; Avari, M.; Cherian, B.P.; Suarez, C. COVID-19-associated Staphylococcus aureus cavitating pneumonia. BMJ Case Rep. 2021, 14, e243726. [Google Scholar] [CrossRef]
- Cusumano, J.A.; Dupper, A.C.; Malik, Y.; Gavioli, E.M.; Banga, J.; Caban, A.B.; Nadkarni, D.; Obla, A.; Vasa, C.V.; Altman, D.R. Staphylococcus aureus Bacteremia in Patients Infected With COVID-19: A Case Series. Open Forum Infect. Dis. 2019, 7, ofaa518. [Google Scholar] [CrossRef]
- La Vecchia, A.; Ippolito, G.; Taccani, V.; Gatti, E.; Bono, P.; Bettocchi, S.; Pinzani, R.; Tagliabue, C.; Bosis, S.; Marchisio, P. Epidemiology and antimicrobial susceptibility of Staphylococcus aureus in children in a tertiary care pediatric hospital in Milan, Italy, 2017–2021. Ital. J. Pediatr. 2022, 48, 67. [Google Scholar] [CrossRef] [PubMed]
- Arikan, K.; Karadag-Oncel, E.; Aycan, E.; Sancak, A.; Ceyhan, M. Molecular characteristics of Staphyloccus aureus strains isolated from nasal samples of sixth year medical students during their pediatric services practices. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Aires-de-Sousa, M. Methicillin-resistant Staphylococcus aureus among animals: Current overview. Clin. Microbiol. Infect. 2017, 23, 373–380. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, H.; Wang, B.; Zhou, Y.; Xu, Y.; Rao, L.; Ai, W.; Guo, Y.; Wu, X.; Yu, J.; et al. Identification of methicillin resistant Staphylococcus aureus ST8 isolates in China with potential high virulence. Emerg. Microbes Infect. 2022, 11, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Chuang, Y.C.; Wang, J.T.; Sheng, W.H.; Chen, C.C.; Chang, S.C. Sequence type 8 as an emerging clone of methicillin-resistant Staphylococcus aureus. Causing bloodstream infections in Taiwan. Emerg. Microbes Infect. 2021, 10, 1908–1918. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Fang, T.H.; Wu, T.H.; Chang, Y.F.; Sung, C.H. Sequence types 8, 59, and 45 methicillin resistant Staphylococcus aureus as the predominant strains causing skin and soft tissue infections in Taiwan’s prisons and jails. J. Microbiol. Immunol. Infect. 2022, 55, 1239–1245. [Google Scholar] [CrossRef]
- Negrete-González, C.; Turrubiates-Martínez, E.; Galicia-Cruz, O.G.; Noyola, D.E.; Martínez-Aguilar, G.; Pérez-González, L.F.; González-Amaro, R.; Niño-Moreno, P. High prevalence of t895 and t9364 spa types of methicillin-resistant Staphylococcus aureus in a tertiary-care hospital in Mexico: Different lineages of clonal complex 5. BMC Microbiol. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Baquero, F.; Corander, J. Global Emergence and Dissemination of Enterococci as Nosocomial Pathogens: Attack of the Clones? Front. Microbiol. 2016, 7, 788. [Google Scholar] [CrossRef]
- Raza, T.; Ullah, S.R.; Mehmood, K. Andleeb Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 2018, 68, 768–772. [Google Scholar]
- Van Tyne, D.; Gilmore, M.S. Raising the alarmone: Within-host evolution of antibiotic-tolerant Enterococcus faecium. mBio 2017, 8, e00066-17. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Clewell, D.B.; Ike, Y.; Shankar, N. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014; pp. 5–63. [Google Scholar]
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Cormican, M.; Flamm, R.K.; Mendes, R.E.; Jones, R.N. Temporal and Geographic Variation in Antimicrobial Susceptibility and Resistance Patterns of Enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 15, S54–S62. [Google Scholar] [CrossRef] [PubMed]
- Kampmeier, S.; Tönnies, H.; Correa-Martinez, C.L.; Mellmann, A.; Schwierzeck, V. A nosocomial cluster of vancomycin resistant enterococci among COVID-19 patients in an intensive care unit. Antimicrob. Resist. Infect. Control 2020, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Bogdanová, K.; Doubravská, L.; Vágnerová, I.; Hricová, K.; Pudová, V.; Röderová, M.; Papajk, J.; Uvízl, R.; Langová, K.; Kolář, M. Clostridioides difficile and Vancomycin-Resistant Enterococci in COVID-19 Patients with Severe Pneumonia. Life 2021, 11, 1127. [Google Scholar] [CrossRef]
- Bonazzetti, C.; Morena, V.; Giacomelli, A.; Oreni, L.; Casalini, G.; Galimberti, L.R. Unexpectedly high frequency of enterococcal bloodstream infections in coronavirus disease 2019 patients admitted to an Italian ICU: An observational study. Crit. Care Med. 2021, 49, e31–e40. [Google Scholar] [CrossRef]
- Giacobbe, D.R.; Battaglini, D.; Ball, L.; Brunetti, I.; Bruzzone, B.; Codda, G.; Crea, F.; De Maria, A.; Dentone, C.; Di Biagio, A.; et al. Bloodstream infections in critically ill patients with COVID-19. Eur. J. Clin. Investig. 2020, 50, e13319. [Google Scholar] [CrossRef]
- Durán-Manuel, E.M.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Ibáñez-Cervantes, G.; Gaytán-Cervantes, J.; González-Torres, C.; Quiroga-Vargas, E.; Calzada-Mendoza, C.C.; Cureño-Díaz, M.A.; Fernández-Sánchez, V.; et al. Massive sequencing of the V3-V4 hypervariable region of bronchoalveolar lavage from patients with COVID-19 and VAP reveals the collapse of the pulmonary microbiota. J. Med. Microbiol. 2022, 71. [Google Scholar] [CrossRef]
- Porto, A.P.M.; Borges, I.C.; Buss, L.; Machado, A.; Bassetti, B.R.; Cocentino, B.; Bicalho, C.S.; Carrilho, C.; Rodrigues, C.; Neto, E.A.S.; et al. HAI/COVID-19 Brazilian task force. Healthcare-associated infections on the ICU in 21 Brazilian hospitals during the early months of the COVID-19 pandemic: An ecological study. Infect. Control Hosp. Epidemiol. 2022, 44, 284–290. [Google Scholar] [CrossRef]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standard Institute (CLSI). Performance Standards for Anti-Microbial Susceptibility Testing, 31st ed.; CLSI Supplement M100; Clinical and Laboratory Standard Institute (CLSI): Wayne, PA, USA, 2021; ISBN 978-1-68440-105-5. [Google Scholar]
- Gilmore, M.S.; Lebreton, F.; van Schaik, W. Genomic transition of enterococci from gut commensalsto leading causes of multidrugresistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 2013, 16, 10–16. [Google Scholar] [CrossRef]
- Rice, L.B.; Marshall, S.H. Evidence of incorporation of the chromosomal B-lactamase gene of Enterococcus faecalis CH19 into a transposon derived from staphylococci. Antimicrob. Agents Chemother. 1992, 36, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.; Le Bouguénec, C.; Gutmann, L.; Horaud, T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 1985, 131, 1933–1940. [Google Scholar] [CrossRef] [PubMed]
- Shlaes, D.M.; Al-Obeid, S.; Shlaes, J.H.; Boisivon, A.; Williamson, R. Inducible, transferable resistance to vancomycin in Enterococcus faecium, D399. J. Antimicrob. Chemother. 1989, 23, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Arthur, M.; Courvalin, P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 1993, 37, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Toc, D.A.; Butiuc-Keul, A.L.; Iordache, D.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Chiorean, C.; Neagoe, D.S.; Gheorghiu, L.; et al. Descriptive Analysis of Circulating Antimicrobial Resistance Genes in Vancomycin-Resistant Enterococcus (VRE) during the COVID-19 Pandemic. Biomedicines 2022, 10, 1122. [Google Scholar] [CrossRef]
- Kanematsu, E.; Deguchi, T.; Yasuda, M.; Kawamura, T.; Nishino, Y.; Kawada, Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 1998, 42, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef]
- Vester, B. The cfr and cfr-like multiple resistance genes. Res. Microbiol. 2018, 169, 61–66. [Google Scholar] [CrossRef]
- Miller, W.R.; Bayer, A.S.; Arias, C.A. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harb. Perspect. Med. 2016, 6, 026997. [Google Scholar] [CrossRef]
- Homan, W.L.; Tribe, D.; Poznanski, S.; Li, M.; Hogg, G.; Spalburg, E.; van Embden, J.D.A.; Willems, R.J.L. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002, 40, 1963–1971. [Google Scholar] [CrossRef]
- Kuch, A.; Willems, R.J.L.; Werner, G.; Coque, T.M.; Hammerum, A.M.; Sundsfjord, A.; Klare, I.; Ruiz-Garbajosa, P.; Simonsen, G.S.; van Luit-Asbroek, M.; et al. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. J. Antimicrob. Chemother. 2012, 67, 551–558. [Google Scholar] [CrossRef] [PubMed]
| Infection | Asia-Pacific | Europe | Latin America | North America |
|---|---|---|---|---|
| Pneumonias | 54.6% | 41.2% | 38.5% | 41.4% |
| Blood | 25.5% | 39.8% | 46% | 33.3% |
| Resistance Mechanism | Family/Type |
|---|---|
| β-lactamases | TEM, SHV, CTX, KPC, GES, IMP, NDM, KPC, AmpC OXA-10, OXA-30, OXA-48, OXA-181 |
| Aminoglycoside-modifying enzymes | Aminoglycoside acetyl-transferases, aminoglycoside adenylyl-transferases, and aminoglycoside phosphotransferases |
| Target site modifications | GyrA (Ala67-Gln106); GyrB (Asp426-Lys447); GyrA (Ala67-Gln106); GyrB (Asp426-Lys447) qnr (pentapeptide proteins, families A, B, C, D, S, and VC), mcr genes, Operons arn, pbg, pmrCAB, crrAB Mutations in pmrA, pmrB, pmrC, pmrD, crrA, ramA, opmW, mgrB |
| Permeability defects | Mutations in ompK35 and ompK36, Efflux Pumps (AcrAB-TolC and OqxAB) |
| CA-MRSA | HA-MRSA | |
|---|---|---|
| Antimicrobial susceptibility profiling | They do not commonly exhibit resistance to beta-lactam antibiotics. | They are usually resistant to several classes of non-beta-lactam antibiotics. |
| Genetic characteristics | Have a smaller SCCmec sequence of type III, IV, or V | They host large SCCmec type I, II, III, or IV elements. |
| Virulence factors | The pvl gene coding for leukocidin toxin is predominant. Virulence genes coding for haemolysins and toxin-exposing superantigens are expressed at high levels. | The pvl gene is occasionally found. Decreased expression of virulence genes encoding for haemolysins and toxin-exposing superantigens. |
| Source of infection | Related to skin and soft tissue. Dangerous and fulminant infections, with further clinical complications. | They are more invasive, not only related to skin and soft tissue. |
| Resistance Mechanism | Gene(s)/Operon(s) |
|---|---|
| Aminoglycoside-modifying enzymes | aac-2 -aph-2”-le, aph-3 -IIIa |
| Acetylation of chloramphenicol | Cat |
| Permeability defects | lsa(A), tet(L |
| Alteration in membrane charge and fluidity | liaFSR |
| Ribosomal methylation | ermB, cfr |
| Target site modifications | gyrA, parC, vanA, vanB, vanD, vanM, rpoB, pbp4 (E. faecalis), pbp5 (E. faecium) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loyola-Cruz, M.Á.; Gonzalez-Avila, L.U.; Martínez-Trejo, A.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Bello-López, J.M.; Castro-Escarpulli, G. ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic. Pathogens 2023, 12, 743. https://doi.org/10.3390/pathogens12050743
Loyola-Cruz MÁ, Gonzalez-Avila LU, Martínez-Trejo A, Saldaña-Padilla A, Hernández-Cortez C, Bello-López JM, Castro-Escarpulli G. ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic. Pathogens. 2023; 12(5):743. https://doi.org/10.3390/pathogens12050743
Chicago/Turabian StyleLoyola-Cruz, Miguel Ángel, Luis Uriel Gonzalez-Avila, Arturo Martínez-Trejo, Andres Saldaña-Padilla, Cecilia Hernández-Cortez, Juan Manuel Bello-López, and Graciela Castro-Escarpulli. 2023. "ESKAPE and Beyond: The Burden of Coinfections in the COVID-19 Pandemic" Pathogens 12, no. 5: 743. https://doi.org/10.3390/pathogens12050743