Effect of Gravity on Bacterial Adhesion to Heterogeneous Surfaces
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Gravitational Sedimentation Affects the Concentration of Bacteria near the Lower Surface
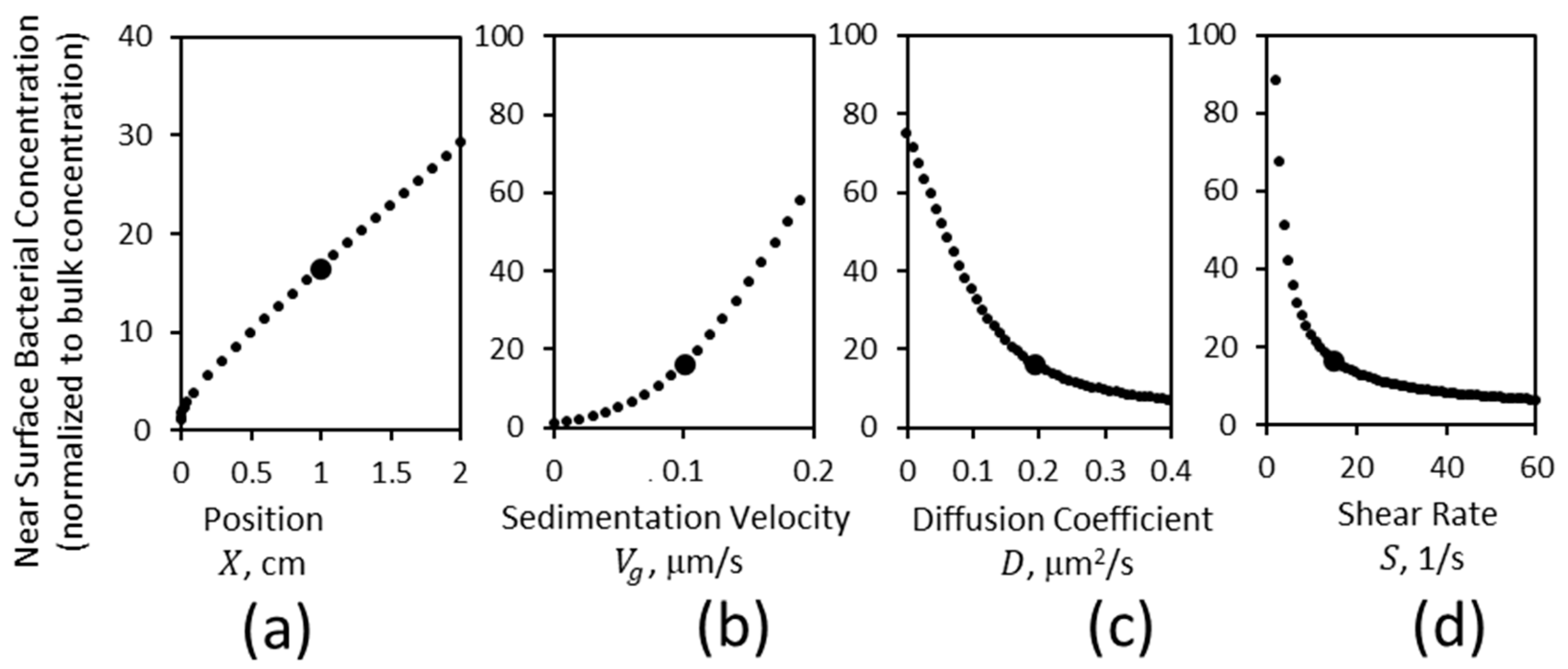
3.2. Effect of Mass Transport on the Concentration of Bacteria near a Lower Surface
3.3. Effect of Mass Transport on Bacterial Adhesion
3.4. Sensitivity of Bacterial Adhesion to Binding Kinetics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karlowsky, J.A.; Kelly, L.J.; Thornsberry, C.; Jones, M.E.; Sahm, D.F. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob. Agents Chemother. 2002, 46, 2540–2545. [Google Scholar] [CrossRef] [Green Version]
- Poutanen, S.M.; de Azavedo, J.; Willey, B.M.; Low, D.E.; MacDonald, K.S. Molecular characterization of multidrug resistance in Streptococcus mitis. Antimicrob. Agents Chemother. 1999, 43, 1505–1507. [Google Scholar] [CrossRef] [Green Version]
- Hsu, R.B.; Lin, F.Y. Effect of penicillin resistance on presentation and outcome of nonenterococcal streptococcal infective endocarditis. Cardiology 2006, 105, 234–239. [Google Scholar] [CrossRef]
- Schito, G.C.; Naber, K.G.; Botto, H.; Palou, J.; Mazzei, T.; Gualco, L.; Marchese, A. The ARESC study: An international survey on the antimicrobial resistance of pathogens involved in uncomplicated urinary tract infections. Int. J. Antimicrob. Agents 2009, 34, 407–413. [Google Scholar] [CrossRef] [Green Version]
- Zhanel, G.G.; Hisanaga, T.L.; Laing, N.M.; DeCorby, M.R.; Nichol, K.A.; Palatnick, L.P.; Johnson, J.; Noreddin, A.; Harding, G.K.; Nicolle, L.E.; et al. Antibiotic resistance in outpatient urinary isolates: Final results from the North American Urinary Tract Infection Collaborative Alliance (NAUTICA). Int. J. Antimicrob. Agents 2005, 26, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Culshaw, S.; Jones, B.; Williams, C. Are we any closer to beating the biofilm: Novel methods of biofilm control. Curr. Opin. Infect. Dis. 2010, 23, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Ofek, I.; Hasty, D.L.; Sharon, N. Anti-adhesion therapy of bacterial diseases: Prospects and problems. FEMS Immunol. Med. Microbiol. 2003, 38, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Lindhorst, T.K. The Bacterial Lectin FimH, a Target for Drug Discovery—Carbohydrate Inhibitors of Type 1 Fimbriae-Mediated Bacterial Adhesion. Eur. J. Org. Chem. 2011, 2011, 3583–3609. [Google Scholar] [CrossRef]
- Langermann, S.; Möllby, R.; Burlein, J.E.; Palaszynski, S.R.; Auguste, C.G.; DeFusco, A.; Strouse, R.; Schenerman, M.A.; Hultgren, S.J.; Pinkner, J.S.; et al. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 2000, 181, 774–778. [Google Scholar] [CrossRef] [Green Version]
- Duddridge, J.E.; Kent, C.A.; Laws, J.F. Effect of surface shear stress on the attachment of Pseudomonas fluorescens to stainless steel under defined flow conditions. Biotechnol. Bioeng. 1982, 24, 153–164. [Google Scholar] [CrossRef]
- Pratt-Terpstra, I.H.; Weerkamp, A.H.; Busscher, H.J. Adhesion of oral streptococci from a flowing suspension to uncoated and albumin-coated surfaces. J. Gen. Microbiol. 1987, 133 Pt 11, 3199–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christersson, C.E.; Glantz, P.O.; Baier, R.E. Role of temperature and shear forces on microbial detachment. Eur. J. Oral Sci. 1988, 96, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.W.; Anderson, J.M.; Marchant, R.E. Platelet-mediated adhesion of Staphylococcus epidermidis to hydrophobic NHLBI reference polyethylene. J. Biomed. Mater. Res. 1993, 27, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, R.B.; Cooper, S.L. Analysis of Shear-Dependent Bacterial Adhesion Kinetics to Biomaterial Surfaces. Aiche J. 1995, 41, 2160–2174. [Google Scholar] [CrossRef]
- Wang, I.W.; Anderson, J.M.; Jacobs, M.R.; Marchant, R.E. Adhesion of Staphylococcus epidermidis to biomedical polymers: Contributions of surface thermodynamics and hemodynamic shear conditions. J. Biomed. Mater. Res. 1995, 29, 485–493. [Google Scholar] [CrossRef]
- Higashi, J.M.; Wang, I.W.; Shlaes, D.M.; Anderson, J.M.; Marchant, R.E. Adhesion of Staphylococcus epidermidis and transposon mutant strains to hydrophobic polyethylene. J. Biomed. Mater. Res. 1998, 39, 341–350. [Google Scholar] [CrossRef]
- Mohamed, N.; Teeters, M.A.; Patti, J.M.; Hook, M.; Ross, J.M. Inhibition of Staphylococcus aureus adherence to collagen under dynamic conditions. Infect. Immun. 1999, 67, 589–594. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.J.; Mohamed, N.; Ross, J.M. Shear stress affects the kinetics of Staphylococcus aureus adhesion to collagen. Biotechnol. Prog. 2000, 16, 1086–1090. [Google Scholar] [CrossRef]
- Reddy, K.; Ross, J.M. Shear stress prevents fibronectin binding protein-mediated Staphylococcus aureus adhesion to resting endothelial cells. Infect. Immun. 2001, 69, 3472–3475. [Google Scholar] [CrossRef] [Green Version]
- Roosjen, A.; Boks, N.P.; van der Mei, H.C.; Busscher, H.J.; Norde, W. Influence of shear on microbial adhesion to PEO-brushes and glass by convective-diffusion and sedimentation in a parallel plate flow chamber. Colloids Surf. B: Biointerfaces 2005, 46, 1–6. [Google Scholar] [CrossRef]
- Azevedo, N.F.; Pinto, A.R.; Reis, N.M.; Vieira, M.J.; Keevil, C.W. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl. Environ. Microbiol. 2006, 72, 2936–2941. [Google Scholar] [CrossRef] [Green Version]
- Boks, N.P.; Norde, W.; van der Mei, H.C.; Busscher, H.J. Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology 2008, 154, 3122–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsikogianni, M.G.; Missirlis, Y.F. Bacterial adhesion onto materials with specific surface chemistries under flow conditions. J. Mater. Sci. Mater. Med. 2010, 21, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Weaver, W.M.; Dharmaraja, S.; Milisavljevic, V.; Di Carlo, D. The effects of shear stress on isolated receptor-ligand interactions of Staphylococcus epidermidis and human plasma fibrinogen using molecularly patterned microfluidics. Lab Chip 2011, 11, 883–889. [Google Scholar] [CrossRef]
- Arpa-Sancet, M.P.; Christophis, C.; Rosenhahn, A. Microfluidic assay to quantify the adhesion of marine bacteria. Biointerphases 2012, 7, 26. [Google Scholar] [CrossRef]
- Xu, L.C.; Siedlecki, C.A. Submicron-textured biomaterial surface reduces staphylococcal bacterial adhesion and biofilm formation. Acta Biomater. 2012, 8, 72–81. [Google Scholar] [CrossRef]
- Sharma, S.; Jaimes-Lizcano, Y.A.; McLay, R.B.; Cirino, P.C.; Conrad, J.C. Subnanometric Roughness Affects the Deposition and Mobile Adhesion of Escherichia coli on Silanized Glass Surfaces. Langmuir 2016, 32, 5422–5433. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokova, V.; McVeigh, A.L.; Kidd, B.; Yakovenko, O.; Thomas, W.E.; Sokurenko, E.V.; Savarino, S.J. Shear-enhanced binding of intestinal colonization factor antigen I of enterotoxigenic Escherichia coli. Mol. Microbiol. 2010, 76, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Thomas, W.E.; Nilsson, L.; Forero, M.; Sokurenko, E.V.; Vogel, V. Shear-dependent “stick-and-roll” adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 2004, 53, 1545–1557. [Google Scholar] [CrossRef]
- Missirlis, Y.F.; Katsikogianni, M. Theoretical and experimental approaches of bacteria-biomaterial interactions. Materwiss Werksttech 2007, 38, 983–994. [Google Scholar] [CrossRef]
- Nejadnik, M.R.; van der Mei, H.C.; Busscher, H.J.; Norde, W. Determination of the shear force at the balance between bacterial attachment and detachment in weak-adherence systems, using a flow displacement chamber. Appl. Environ. Microbiol. 2008, 74, 916–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascari, L.; Ymele-Leki, P.; Eggleton, C.D.; Speziale, P.; Ross, J.M. Fluid shear contributions to bacteria cell detachment initiated by a monoclonal antibody. Biotechnol. Bioeng. 2003, 83, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, L.M.; Thomas, W.E.; Sokurenko, E.V.; Vogel, V. Elevated shear stress protects Escherichia coli cells adhering to surfaces via catch bonds from detachment by soluble inhibitors. Appl. Environ. Microbiol. 2006, 72, 3005–3010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kisiela, D.I.; Rodriguez, V.B.; Tchesnokova, V.; Avagyan, H.; Aprikian, P.; Liu, Y.; Wu, X.-R.; Thomas, W.E.; Sokurenko, E.V. Conformational inactivation induces immunogenicity of the receptor-binding pocket of a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2013, 110, 19089–19094. [Google Scholar] [CrossRef] [PubMed]
- Kisiela, D.I.; Avagyan, H.; Friend, D.; Jalan, A.; Gupta, S.; Interlandi, G.; Liu, Y.; Tchesnokova, V.; Rodriguez, V.B.; Sumida, J.P.; et al. Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic E coli. PLoS Pathog. 2015, 11, e1004857. [Google Scholar] [CrossRef] [Green Version]
- Tchesnokova, V.; Aprikian, P.; Kisiela, D.; Gowey, S.; Korotkova, N.; Thomas, W.; Sokurenko, E. Type 1 fimbrial adhesin FimH elicits an immune response that enhances cell adhesion of Escherichia coli. Infect. Immun. 2011, 79, 3895–3904. [Google Scholar] [CrossRef] [Green Version]
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. 2014, 28, 1–13. [Google Scholar] [CrossRef]
- Klein, R.D.; Hultgren, S.J. Urinary tract infections: Microbial pathogenesis, host–pathogen interactions and new treatment strategies. Nat. Rev. Microbiol. 2020, 18, 211–226. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Diekema, D.J.; Beekmann, S.E.; Chapin, K.C.; Morel, K.A.; Munson, E.; Doern, G.V. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J. Clin. Microbiol. 2003, 41, 3655–3660. [Google Scholar] [CrossRef] [Green Version]
- Laupland, K.B.; Church, D.L. Population-based epidemiology and microbiology of community-onset bloodstream infections. Clin. Microbiol. Rev. 2014, 27, 647–664. [Google Scholar] [CrossRef] [Green Version]
- Harles, M.P.; Kali, A.; Easow, J.M.; Joseph, N.M.; Ravishankar, M.; Srinivasan, S.; Kumar, S.; Umadevi, S. Ventilator-associated pneumonia. Australas. Med. J. 2014, 7, 334. [Google Scholar] [CrossRef]
- Dale, A.P.; Woodford, N. Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. J. Infection. 2015, 71, 615–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, W.E.; Trintchina, E.; Forero, M.; Vogel, V.; Sokurenko, E.V. Bacterial adhesion to target cells enhanced by shear force. Cell 2002, 109, 913–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aprikian, P.; Interlandi, G.; Kidd, B.A.; Le Trong, I.; Tchesnokova, V.; Yakovenko, O.; Whitfield, M.J.; Bullitt, E.; Stenkamp, R.E.; Thomas, W.E.; et al. The bacterial fimbrial tip acts as a mechanical force sensor. PLoS Biol. 2011, 9, e1000617. [Google Scholar] [CrossRef] [PubMed]
- Le Trong, I.; Aprikian, P.; Kidd, B.A.; Forero-Shelton, M.; Tchesnokova, V.; Rajagopal, P.; Rodriguez, V.; Interlandi, G.; Klevit, R.; Vogel, V.; et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell 2010, 141, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Sokurenko, E.V.; Tchesnokova, V.; Interlandi, G.; Klevit, R.; Thomas, W.E. Neutralizing Antibodies Against Allosteric Proteins: Insights From a Bacterial Adhesin. J. Mol. Biol. 2022, 434, 167717. [Google Scholar] [CrossRef]
- Mohamed, N.; Visai, L.; Speziale, P.; Ross, J.M. Quantification of Staphylococcus aureus cell surface adhesins using flow cytometry. Microb. Pathog. 2000, 29, 357–361. [Google Scholar] [CrossRef]
- Fallgren, C.; Ljungh, A.; Shenkman, B.; Varon, D.; Savion, N. Venous shear stress enhances platelet mediated staphylococcal adhesion to artificial and damaged biological surfaces. Biomaterials 2002, 23, 4581–4589. [Google Scholar] [CrossRef]
- Domka, J.; Lee, J.; Wood, T.K. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 2006, 72, 2449–2459. [Google Scholar] [CrossRef] [Green Version]
- Raya, A.; Sodagari, M.; Pinzon, N.M.; He, X.; Newby, B.M.Z.; Ju, L.K. Effects of rhamnolipids and shear on initial attachment of Pseudomonas aeruginosa PAO1 in glass flow chambers. Environ. Sci. Pollut. Res. 2010, 17, 1529–1538. [Google Scholar] [CrossRef]
- Lecuyer, S.; Rusconi, R.; Shen, Y.; Forsyth, A.; Vlamakis, H.; Kolter, R.; Stone, H.A. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys. J. 2011, 100, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Claes, J.; Vanassche, T.; Peetermans, M.; Liesenborghs, L.; Vandenbriele, C.; Vanhoorelbeke, K.; Missiakas, D.; Schneewind, O.; Hoylaerts, M.F.; Heying, R.; et al. Adhesion of Staphylococcus aureus to the vessel wall under flow is mediated by von Willebrand factor-binding protein. Blood 2014, 124, 1669–1676. [Google Scholar] [CrossRef]
- Liesenborghs, L.; Peetermans, M.; Claes, J.; Veloso, T.R.; Vandenbriele, C.; Criel, M.; Lox, M.; Peetermans, W.E.; Heilbronner, S.; de Groot, P.G.; et al. Shear-Resistant Binding to von Willebrand Factor Allows Staphylococcus lugdunensis to Adhere to the Cardiac Valves and Initiate Endocarditis. J. Infect. Dis. 2016, 213, 1148–1156. [Google Scholar] [CrossRef] [Green Version]
- Yakovenko, O.; Nunez, J.; Bensing, B.; Yu, H.; Mount, J.; Zeng, J.; Hawkins, J.; Chen, X.; Sullam, P.M.; Thomas, W. Serine-Rich Repeat Adhesins Mediate Shear-Enhanced Streptococcal Binding to Platelets. Infect. Immun. 2018, 86, e00160-18. [Google Scholar] [CrossRef] [Green Version]
- Kisiela, D.I.; Kramer, J.J.; Tchesnokova, V.; Aprikian, P.; Yarov-Yarovoy, V.; Clegg, S.; Sokurenko, E.V. Allosteric catch bond properties of the FimH adhesin from Salmonella enterica serovar Typhimurium. J. Biol. Chem. 2011, 286, 38136–38147. [Google Scholar] [CrossRef] [Green Version]
- Ding, A.M.; Palmer, R.J., Jr.; Cisar, J.O.; Kolenbrander, P.E. Shear-enhanced oral microbial adhesion. Appl. Environ. Microbiol. 2010, 76, 1294–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriarty, T.J.; Shi, M.; Lin, Y.-P.; Ebady, R.; Zhou, H.; Odisho, T.; Hardy, P.-O.; Salman-Dilgimen, A.; Wu, J.; Weening, E.H.; et al. Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol. Microbiol. 2012, 86, 1116–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niddam, A.F.; Ebady, R.; Bansal, A.; Koehler, A.; Hinz, B.; Moriarty, T.J. Plasma fibronectin stabilizes Borrelia burgdorferi-endothelial interactions under vascular shear stress by a catch-bond mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, E3490–E3498. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W. Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 2008, 10, 39–57. [Google Scholar] [CrossRef] [Green Version]
- Sokurenko, E.V.; Vogel, V.; Thomas, W.E. Catch-bond mechanism of force-enhanced adhesion: Counterintuitive, elusive, but... widespread? Cell Host Microbe 2008, 4, 314–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busscher, H.J.; van der Mei, H.C. Microbial adhesion in flow displacement systems. Clin. Microbiol. Rev. 2006, 19, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Busscher, H.J.; Norde, W.; Sjollema, J. Analysis of the contribution of sedimentation to bacterial mass transport in a parallel plate flow chamber. Colloids Surf. B Biointerfaces 2011, 84, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Padron, G.C.; Shuppara, A.M.; Palalay, J.J.S.; Sharma, A.; Sanfilippo, J.E. Bacteria in Fluid Flow. J. Bacteriol. 2023, 205, e00400–e00422. [Google Scholar] [CrossRef]
- Myszka, D.G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997, 8, 50–57. [Google Scholar] [CrossRef]
- Myszka, D.G.; He, X.; Dembo, M.; Morton, T.A.; Goldstein, B. Extending the range of rate constants available from BIACORE: Interpreting mass transport-influenced binding data. Biophys. J. 1998, 75, 583–594. [Google Scholar] [CrossRef] [Green Version]
- Rich, R.L.; Cannon, M.J.; Jenkins, J.; Pandian, P.; Sundaram, S.; Magyar, R.; Brockman, J.; Lambert, J.; Myszka, D.G. Extracting kinetic rate constants from surface plasmon resonance array systems. Anal. Biochem. 2008, 373, 112–120. [Google Scholar] [CrossRef]
- Korber, D.R.; Lawrence, J.R.; Zhang, L.; Caldwell, D.E. Effect of gravity on bacterial deposition and orientation in laminar flow environments. Biofouling 1990, 2, 335–350. [Google Scholar] [CrossRef]
- Munn, L.L.; Melder, R.J.; Jain, R.K. Analysis of cell flux in the parallel plate flow chamber: Implications for cell capture studies. Biophys. J. 1994, 67, 889–895. [Google Scholar] [CrossRef] [Green Version]
- Yakovenko, O.; Tchesnokova, V.; Sokurenko, E.V.; Thomas, W.E. Inactive conformation enhances binding function in physiological conditions. Proc. Natl. Acad. Sci. USA 2015, 112, 9884–9889. [Google Scholar] [CrossRef]
- Blanchard, B.; Nurisso, A.; Hollville, E.; Tétaud, C.; Wiels, J.; Pokorná, M.; Wimmerová, M.; Varrot, A.; Imberty, A. Structural basis of the preferential binding for globo-series glycosphingolipids displayed by Pseudomonas aeruginosa lectin I. J. Mol. Biol. 2008, 383, 837–853. [Google Scholar] [CrossRef] [PubMed]
- Padler-Karavani, V.; Song, X.; Yu, H.; Hurtado-Ziola, N.; Huang, S.; Muthana, S.; Chokhawala, H.A.; Cheng, J.; Verhagen, A.; Langereis, M.A.; et al. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J. Biol. Chem. 2012, 287, 22593–22608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Kerpel, M.; Van Molle, I.; Brys, L.; Wyns, L.; De Greve, H.; Bouckaert, J. N-terminal truncation enables crystallization of the receptor-binding domain of the FedF bacterial adhesin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62 Pt 12, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.L.; Pilobello, K.T.; Mahal, L.K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2006, 2, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Mader, A.; Gruber, K.; Castelli, R.; Hermann, B.A.; Seeberger, P.H.; Rädler, J.O.; Leisner, M. Discrimination of Escherichia coli strains using glycan cantilever array sensors. Nano Lett. 2012, 12, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Plummer, T.; Hirs, C.; Tench, A.L. The isolation of ribonuclease B, a glycoprotein, from bovine pancreatic juice. J. Biol. Chem. 1963, 238, 1396–1401. [Google Scholar] [CrossRef]
- Johnson, K.C.; Clemmens, E.; Mahmoud, H.; Kirkpatrick, R.; Vizcarra, J.C.; Thomas, W.E. A multiplexed magnetic tweezer with precision particle tracking and bi-directional force control. J. Biol. Eng. 2017, 11, 47. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, L.M.; Thomas, W.E.; Trintchina, E.; Vogel, V.; Sokurenko, E.V. Catch bond-mediated adhesion without a shear threshold: Trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J. Biol. Chem. 2006, 281, 16656–16663. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, M.; Ghose, T.; Thomas, W. Shear-stabilized rolling behavior of E. coli examined with simulations. Biophys. J. 2010, 99, 2470–2478. [Google Scholar] [CrossRef] [Green Version]






| Symbol | Definition | Default Value |
|---|---|---|
| Gravitational sedimentation velocity 1 | ||
| D | Bacterial diffusion coefficient 1 | |
| r | Bacterial radius | 1 µm |
| Effective bacterial association rate 2 | ||
| Initial bacterial concentration 2 | ||
| H | Chamber depth 3 | 254 µm |
| S | Wall shear rate 3 | |
| Location of upstream edge of spot 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hogan, K.; Paul, S.; Lin, G.; Fuerte-Stone, J.; Sokurenko, E.V.; Thomas, W.E. Effect of Gravity on Bacterial Adhesion to Heterogeneous Surfaces. Pathogens 2023, 12, 941. https://doi.org/10.3390/pathogens12070941
Hogan K, Paul S, Lin G, Fuerte-Stone J, Sokurenko EV, Thomas WE. Effect of Gravity on Bacterial Adhesion to Heterogeneous Surfaces. Pathogens. 2023; 12(7):941. https://doi.org/10.3390/pathogens12070941
Chicago/Turabian StyleHogan, Kayla, Sai Paul, Guanyou Lin, Jay Fuerte-Stone, Evgeni V. Sokurenko, and Wendy E. Thomas. 2023. "Effect of Gravity on Bacterial Adhesion to Heterogeneous Surfaces" Pathogens 12, no. 7: 941. https://doi.org/10.3390/pathogens12070941






