The Role of the Nuclear Factor-Kappa B (NF-κB) Pathway in SARS-CoV-2 Infection
Abstract
:1. Introduction
2. COVID-19 Infection and Pathogenesis
3. NF-κB Signaling Pathways
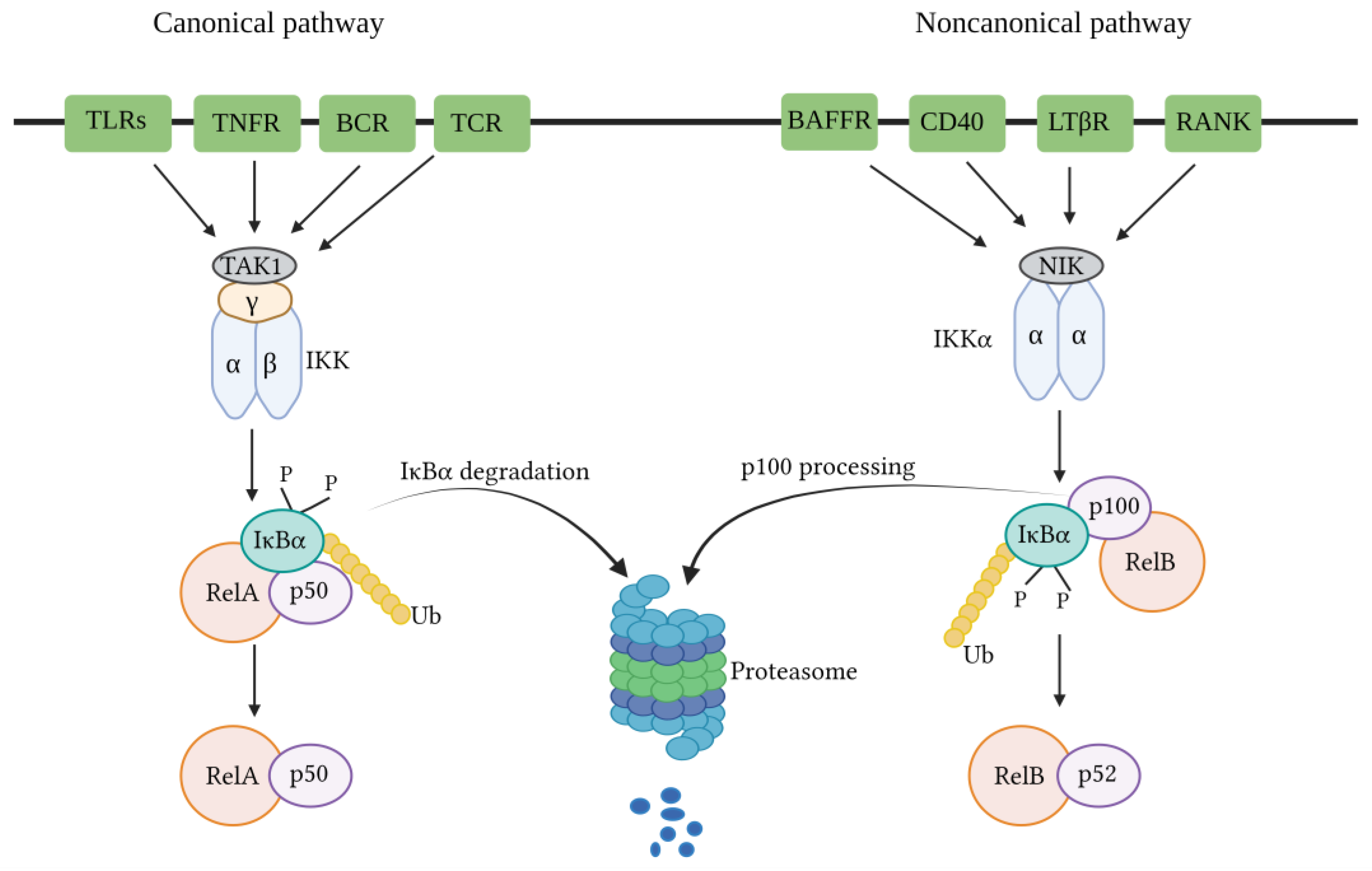
3.1. Canonical NF-κB Pathway
3.2. Non-Canonical NF-κB Pathway
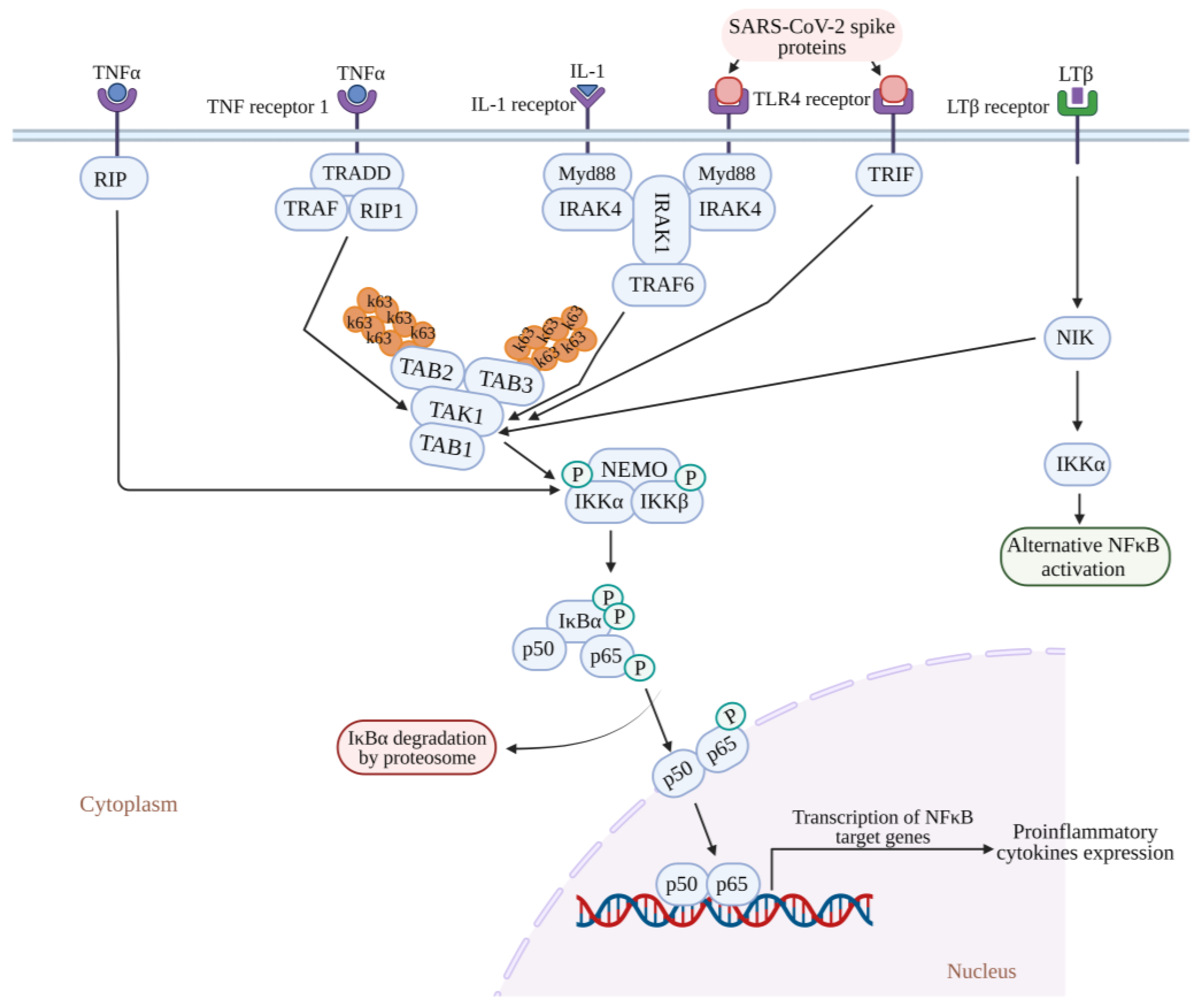
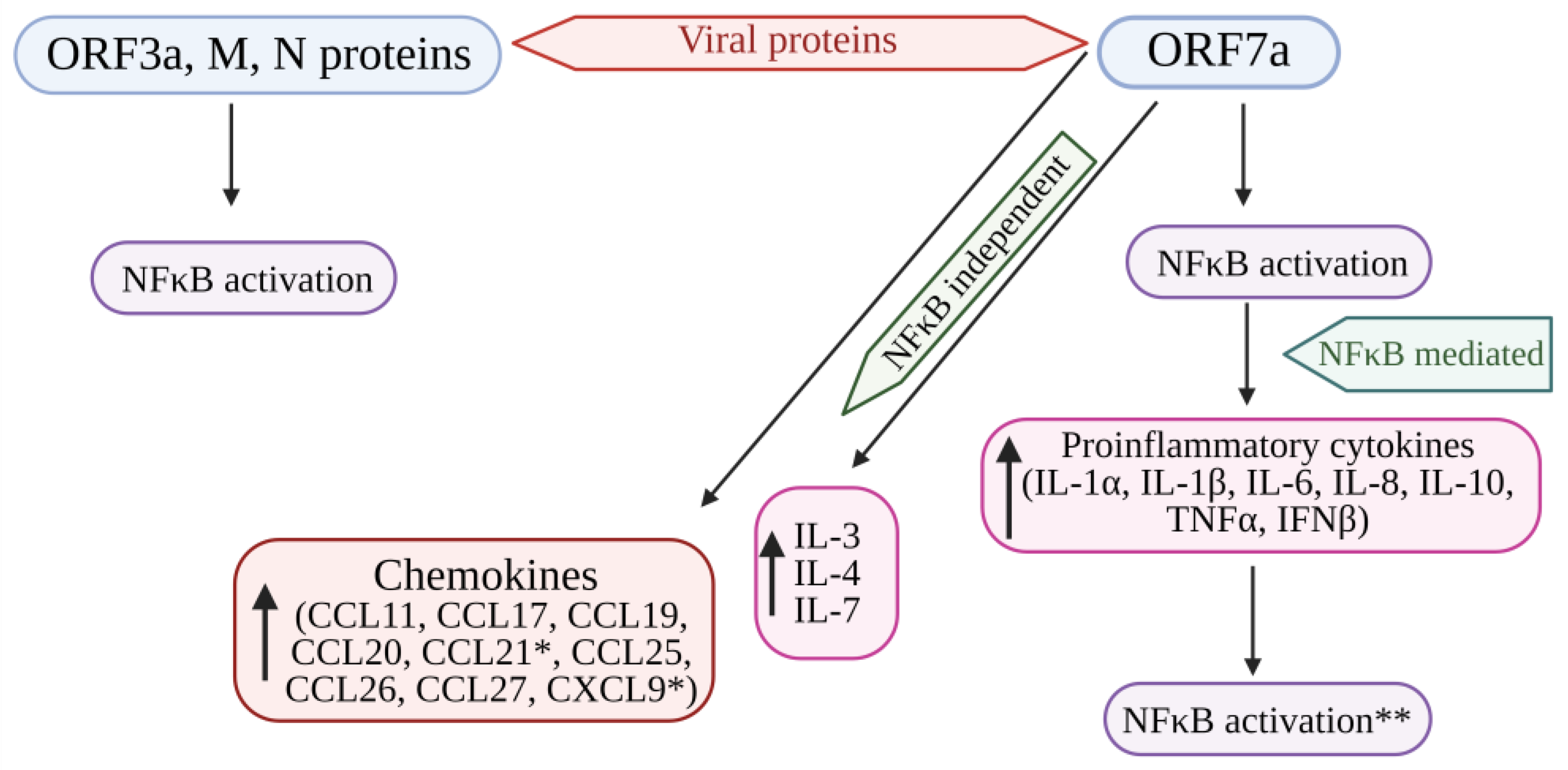
4. NF-κB Signaling in COVID-19 Infection
Recent Knowledge of the Role of NF-kB in COVID-19
5. NF-κB Pathway: A Pharmacological Target in COVID-19
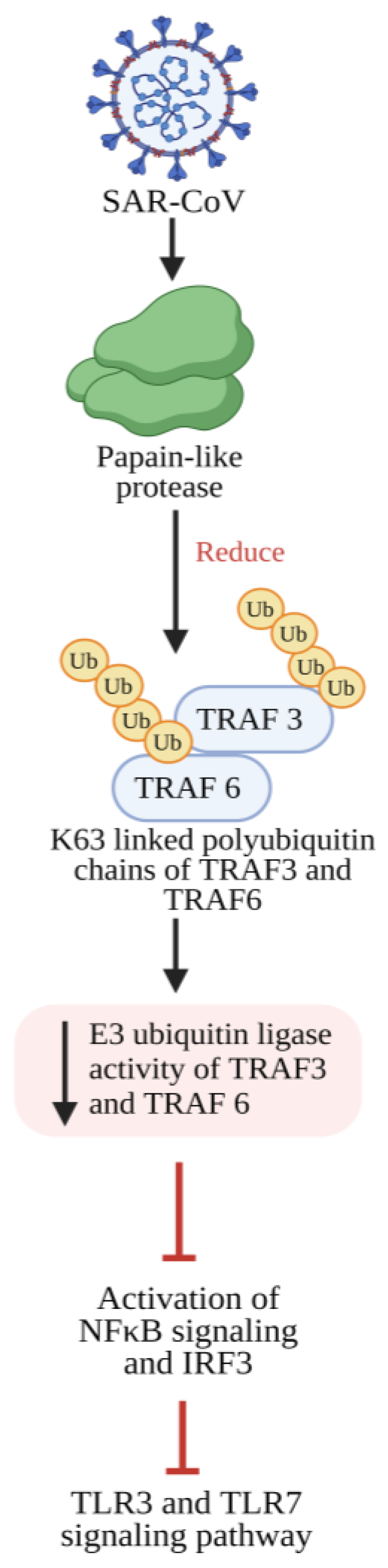
5.1. Inflammatory Response Triggered by the SARS-CoV Spike Protein
5.2. Suppression of NF-κB Activity
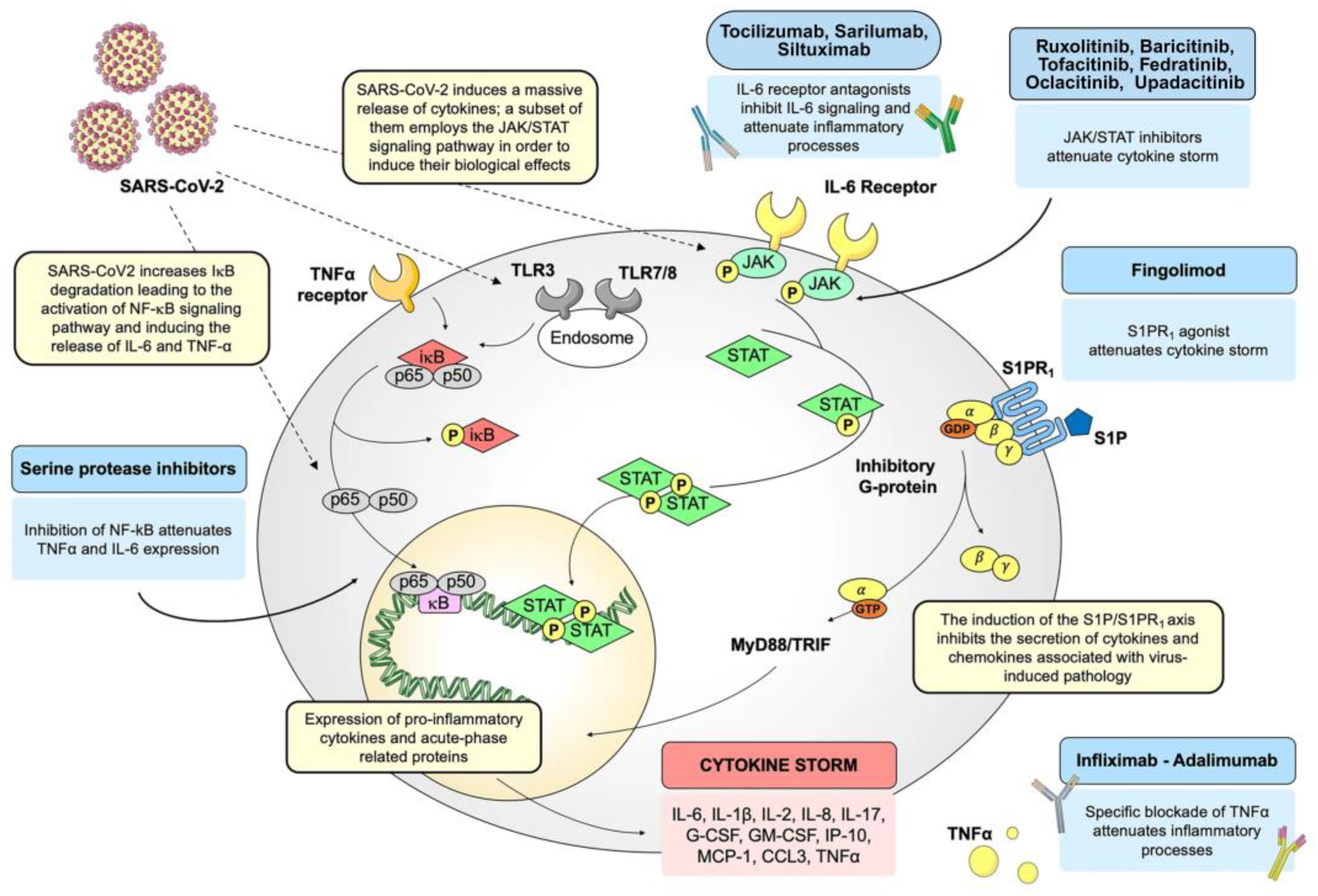
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; John Wherry, E. T cell responses in patients with COVID19. Nat. Rev. Immunol. 2020, 20, 529–536. [Google Scholar] [CrossRef]
- Hui, D.S.; Azhar, I.E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Baron, S.; Fons, M.; Albrecht, T. Viral Pathogenesis. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. Available online: https://www.ncbi.nlm.nih.gov/books/NBK8149/ (accessed on 13 December 2023).
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Bushman, M.; Kishore, N.; Niehus, R.; de Salazar, P.M.; Cowling, B.J.; Lipsitch, M.; Leung, G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat. Med. 2020, 26, 506–510. [Google Scholar] [CrossRef]
- Hassan, S.A.; Sheikh, F.N.; Jamal, S.; Ezeh, J.K.; Akhtar, A. Coronavirus (COVID-19): A Review of Clinical Features, Diagnosis, and Treatment. Cureus 2020, 12, e7355. [Google Scholar] [CrossRef]
- Polidoro, R.B.; Hagan, R.S.; de Santis Santiago, R.; Schmidt, N.W. Overview: Systemic Inflammatory Response Derived from Lung Injury Caused by SARS-CoV-2 Infection Explains Severe Outcomes in COVID-19. Front. Immunol. 2020, 11, 1626. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Nilsson-Payant, B.E.; Uhl, S.; Grimont, A.; Doane, A.S.; Cohen, P.; Patel, R.S.; Higgins, C.A.; Acklin, J.A.; Bram, Y.; Chandar, V.; et al. The NF-kappa B Transcriptional Footprint Is Essential for SARS-CoV-2 Replication. J. Virol. 2021, 95, e0125721. [Google Scholar] [CrossRef]
- Attiq, A.; Yao, L.J.; Afzal, S.; Khan, M.A. The triumvirate of NF-κB, inflammation and cytokine storm in COVID-19. Int. Immunopharmacol. 2021, 101, 108255. [Google Scholar] [CrossRef]
- Li, F.; Zhang, J.; Arfuso, F.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; Kumar, A.P.; Ahn, K.S.; Sethi, G. NF-κB in cancer therapy. Arch. Toxicol. 2015, 89, 711–731. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Cross-talk in NF-κB signaling pathways. Nat. Immunol. 2011, 12, 695. [Google Scholar] [CrossRef]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the War Against Inflammation with Natural Products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef]
- Gerondakis, S.; Grossmann, M.; Nakamura, Y.; Pohl, T.; Grumont, R. Genetic approaches in mice to understand Rel/NF-κB and IκB function: Transgenics and knockouts. Oncogene 1999, 18, 6888–6895. [Google Scholar] [CrossRef]
- Mattson, M.; Meffert, M. Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006, 13, 852–860. [Google Scholar] [CrossRef]
- Courtois, G.; Gilmore, T. Mutations in the NF-κ B signaling pathway: Implications for human disease. Oncogene 2006, 25, 6831–6843. [Google Scholar] [CrossRef]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus- infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar]
- van der Hoek, L.; Pyrc, K.; Berkhout, B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006, 30, 760–773. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Pennisi, M.; Lanza, G.; Falzone, L.; Fisicaro, F.; Ferri, R.; Bella, R. SARS-CoV-2 and the nervous system: From clinical features to molecular mechanisms. Int. J. Mol. Sci. 2020, 21, 5475. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Peng, R.; Wu, L.A.; Wang, Q.; Qi, J.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Cevik, M.; Kuppalli, K.; Kindrachuk, J.; Peiris, M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 2020, 371, m3862. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular Mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020, 94, e02015-19. [Google Scholar] [CrossRef]
- Davidson, A.M.; Wysocki, J.; Batlle, D. Interaction of SARS-CoV-2 and other coronavirus with ACE (Angiotensin-converting enzyme)-2 as their main receptor: Therapeutic implications. Hypertension 2020, 76, 1339–1349. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hofmann-Winkler, H.; Smith, J.C.; Krüger, N.; Arora, P.; Sørensen, L.K.; Søgaard, O.S.; Hasselstrøm, J.B.; Winkler, M.; Hempel, T.; et al. Camostat mesylate inhibits SARS- CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine 2021, 65, 103255. [Google Scholar] [CrossRef]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe COVID-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef]
- Bradley, B.T.; Maioli, H.; Johnston, R.; Chaudhry, I.; Fink, S.L.; Xu, H.; Najafian, B.; Deutsch, G.; Lacy, J.M.; Williams, T.; et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: A case series. Lancet 2020, 396, 320–332. [Google Scholar] [CrossRef]
- Hanley, B.; Naresh, K.N.; Roufosse, C.; Nicholson, A.G.; Weir, J.; Cooke, G.S.; Thursz, M.; Manousou, P.; Corbett, R.; Goldin, R.; et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: A post-mortem study. Lancet Microbe 2020, 1, e245–e253. [Google Scholar] [CrossRef]
- Bhatnagar, J.; Gary, J.; Reagan-Steiner, S.; Estetter, L.B.; Tong, S.; Tao, Y.; Denison, A.M.; Lee, E.; DeLeon-Carnes, M.; Li, Y.; et al. Evidence of severe acute respiratory syndrome coronavirus 2 replication and tropism in the lungs, airways, and vascular endothelium of patients with fatal coronavirus disease 2019: An autopsy case series. J. Infect. Dis. 2021, 223, 752–764. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Li, L.; Liao, H.; Meng, Y.; Li, W.; Han, P.; Liu, K.; Wang, Q.; Li, D.; Zhang, Y.; Wang, L.; et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell 2022, 185, 2952–2960.e10. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef]
- Carcaterra, M.; Caruso, C. Alveolar epithelial cell type II as main target of SARS-CoV-2 virus and COVID-19 development via NF-κB pathway deregulation: A physio-pathological theory. Med. Hypotheses 2021, 146, 110412. [Google Scholar] [CrossRef]
- Fehrenbach, H. Alveolar epithelial type II cell: Defender of the alveolus revisited. Respir. Res. 2001, 2, 33–46. [Google Scholar] [CrossRef]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Zheng, M.; Song, L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020, 17, 536–538. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.F.; Almansa, R.; Menéndez, R.; Mendez, R.; Kelvin, D.J.; Torres, A. Lymphopenic community acquired pneumonia as signature of severe COVID-19 infection. J. Infect. 2020, 80, e23–e24. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- von der Thüsen, J.; van der Eerden, M. Histopathology and genetic susceptibility in COVID-19 pneumonia. Eur. J. Clin. Investig. 2020, 50, e13259. [Google Scholar] [CrossRef]
- Hanley, B.; Lucas, S.B.; Youd, E.; Swift, B.; Osborn, M. Autopsy in suspected COVID-19 cases. J. Clin. Pathol. 2020, 73, 239–242. [Google Scholar] [CrossRef]
- Velavan, T.P.; Meyer, C.G. Mild versus severe COVID-19: Laboratory markers. Int. J. Infect. Dis. 2020, 95, 304–307. [Google Scholar] [CrossRef]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef]
- Binkhorst, M.; Offringa, A.; Hoeven, J. COVID-19: Comprehensive synopsis of suggested pathophysiological mechanisms and repur posed drugs. Preprints 2020, 2020070108. [Google Scholar] [CrossRef]
- Ge, X.; Low, B.; Liang, M.; Fu, J. Angiotensin II directly triggers endo thelial exocytosis via protein kinase C-dependent protein kinase D2 activation. J. Pharmacol. Sci. 2007, 105, 168–176. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Newton, A.H.; Cardani, A.; Braciale, T.J. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin. Immunopathol. 2016, 38, 471–482. [Google Scholar] [CrossRef]
- McGonagle, D.; Sharif, K.; O’Regan, A.; Bridgewood, C. The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun. Rev. 2020, 19, 102537. [Google Scholar] [CrossRef]
- Crayne, C.B.; Albeituni, S.; Nichols, K.E.; Cron, R.Q. The immunology of macrophage activation syndrome. Front. Immunol. 2019, 10, 119. [Google Scholar] [CrossRef]
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 533–535. [Google Scholar] [CrossRef]
- Hamid, T.; Guo, S.Z.; Kingery, J.R.; Xiang, X.; Dawn, B.; Prabhu, S.D. Cardiomyocyte NF-κB RelA promotes adverse remodelling, apoptosis, and endoplasmic reticulum stress in heart failure. Cardiovasc. Res. 2011, 89, 129–138. [Google Scholar] [CrossRef]
- Giridharan, S.; Srinivasan, M. Mechanisms of NF-κB RelA and strategies for therapeutic manipulation. J. Inflamm. Res. 2018, 11, 407–419. [Google Scholar] [CrossRef]
- Albensi, B.C. What Is Nuclear Factor Kappa B (NF-κB) Doing in and to the Mitochondrion? Front. Cell. Dev. Biol. 2019, 7, 154. [Google Scholar] [CrossRef]
- Sun, S.C. Non-canonical NF-κB signaling pathway. Cell Res. 2011, 21, 71–85. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-κ B signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-κ B transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Sun, S.C.; Ley, S.C. New insights into NF-kappa B regulation and function. Trends Immunol. 2008, 29, 469–478. [Google Scholar] [CrossRef]
- Sato, S.; Sanjo, H.; Takeda, K.; Ninomiya-Tsuji, J.; Yamamoto, M.; Kawai, T.; Matsumoto, K.; Takeuchi, O.; Akira, S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005, 6, 1087–1095. [Google Scholar] [CrossRef]
- Shim, J.H.; Xiao, C.; Paschal, A.E.; Bailey, S.T.; Rao, P.; Hayden, M.S.; Lee, K.Y.; Bussey, C.; Steckel, M.; Tanaka, N.; et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005, 19, 2668–2681. [Google Scholar] [CrossRef] [PubMed]
- Solan, N.J.; Miyoshi, H.; Carmona, E.M.; Bren, G.D.; Paya, C.V. RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 2002, 277, 1405–1418. [Google Scholar] [CrossRef]
- Xiao, G.; Harhaj, E.W.; Sun, S.C. NF-κ B-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell. 2001, 7, 401–409. [Google Scholar] [CrossRef]
- Malinin, N.L.; Boldin, M.P.; Kovalenko, A.V.; Wallach, D. MAP3K-related kinase involved in NF-κ B induction by TNF, CD95 and IL-1. Nature 1997, 385, 540–544. [Google Scholar] [CrossRef]
- Senftleben, U.; Cao, Y.; Xiao, G.; Greten, F.R.; Krähn, G.; Bonizzi, G.; Chen, Y.; Hu, Y.; Fong, A.; Sun, S.C.; et al. Activation by IKK alpha of a second, evolutionary conserved, NF-κ B signaling pathway. Science 2001, 293, 1495–1499. [Google Scholar] [CrossRef]
- Claudio, E.; Brown, K.; Park, S.; Wang, H.; Siebenlist, U. BAFF-induced NEMO-independent processing of NF-κ B2 in maturing B cells. Nat. Immunol. 2002, 3, 958–965. [Google Scholar] [CrossRef]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.W.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-κ B pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, M.; Sun, S.C. beta-TrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell Signal. 2006, 18, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Razani, B.; Zarnegar, B.; Ytterberg, A.J.; Shiba, T.; Dempsey, P.W.; Ware, C.F.; Loo, J.A.; Cheng, G. Negative feedback in non-canonical NF-κ B signaling modulates NIK stability through IKK α-mediated phosphorylation. Sci. Signal. 2010, 3, ra41. [Google Scholar] [CrossRef]
- Kandasamy, M. NF-κB signalling as a pharmacological target in COVID-19: Potential roles for IKKβ inhibitors. Naunyn Schmiedebergs Arch. Pharmacol. 2021, 394, 561–567. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 2020, 27, 883–890. [Google Scholar] [CrossRef]
- Hojyo, S.; Uchida, M.; Tanaka, K.; Hasebe, R.; Tanaka, Y.; Murakami, M.; Hirano, T. How COVID-19 induces cytokine storm with high mortality. Infamm. Regen. 2020, 40, 37. [Google Scholar] [CrossRef]
- Huang, K.J.; Su, I.J.; Theron, M.; Wu, Y.C.; Lai, S.K.; Liu, C.C.; Lei, H.Y. An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 2005, 75, 185–194. [Google Scholar] [CrossRef]
- Haque, A.; Hober, D.; Kasper, L.H. Confronting potential infuenza A (H5N1) pandemic with better vaccines. Emerg. Infect. Dis. 2007, 13, 1512–1518. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, Y.; Yang, Z. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016, 13, 3–10. [Google Scholar] [CrossRef]
- Mori, M.; Rothman, A.L.; Kurane, I.; Montoya, J.M.; Nolte, K.B.; Norman, J.E.; Waite, D.C.; Koster, F.T.; Ennis, F.A. High levels of cytokine-producing cells in the lung tissues of patients with fatal hantavirus pulmonary syndrome. J. Infect. Dis. 1999, 179, 295–302. [Google Scholar] [CrossRef]
- Su, C.M.; Wang, L.; Yoo, D. Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021, 11, 13464. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-κ B family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Wan, F.; Lenardo, M.J. Specification of DNA binding activity of NF-κB proteins. Cold Spring Harb. Perspect. Biol. 2009, 1, a000067. [Google Scholar] [CrossRef]
- Song, K.; Li, S. The Role of Ubiquitination in NF-κB Signaling during Virus Infection. Viruses 2021, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-κ B pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Karin, M. How NF-κ B is activated: The role of the Iκ B kinase (IKK) complex. Oncogene 1999, 18, 6867–6874. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Mattson, M.P.; Camandola, S. NF-κB in neuronal plasticity and neurodegenerative disorders. J. Clin. Investig. 2001, 107, 247–254. [Google Scholar] [CrossRef]
- Alcamo, E.; Hacohen, N.; Schulte, L.C.; Rennert, P.D.; Hynes, R.O.; Baltimore, D. Requirement for the NF-κB family member RelA in the development of secondary lymphoid organs. J. Exp. Med. 2002, 195, 233–244. [Google Scholar] [CrossRef]
- Sizemore, N.; Agarwal, A.; Das, K.; Lerner, N.; Sulak, M.; Rani, S.; Ransohoff, R.; Shultz, D.; Stark, G.R. Inhibitor of κB kinase is required to activate a subset of interferon γ-stimulated genes. Proc. Natl. Acad. Sci. USA 2004, 101, 7994–7998. [Google Scholar] [CrossRef]
- Freudlsperger, C.; Bian, Y.; Contag, S.; Burnett, J.; Coupar, J.; Yang, X.; Chen, Z.; Van Waes, C. TGF-β and NF-κB signal pathway cross-talk is mediated through TAK1 and SMAD7 in a subset of head and neck cancers. Oncogene 2013, 32, 1549–1559. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef]
- Schmitz, M.L.; Kracht, M.; Saul, V.V. The intricate interplay between RNA viruses and NF-κB. Biochim. Biophys. Acta 2014, 1843, 2754–2764. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z. Crosstalk of reactive oxygen species and NFκB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef]
- Liao, Q.J.; Ye, L.B.; Timani, K.A.; Zeng, Y.C.; She, Y.L.; Ye, L.; Wu, Z.H. Activation of NF-kB by the full-length nucleocapsid protein of the SARS coronavirus. Acta Biochim. Biophys. Sin 2005, 37, 607–612. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Nieto-Torres, J.L.; Regla-Nava, J.A.; Jimenez-Guardeño, J.M.; Fernandez-Delgado, R.; Fett, C.; Castaño-Rodriguez, C.; Perlman, S.; Enjuanes, L. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014, 88, 913–924. [Google Scholar] [CrossRef]
- Hirano, T.; Murakami, M. COVID-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef]
- Li, S.W.; Wang, C.Y.; Jou, Y.J.; Huang, S.H.; Hsiao, L.H.; Wan, L.; Lin, Y.J.; Kung, S.H.; Lin, C.W. SARS Coronavirus Papain-Like Protease Inhibits the TLR7 Signaling Pathway through Removing Lys63-Linked Polyubiquitination of TRAF3 and TRAF6. Int. J. Mol. Sci. 2016, 17, 678. [Google Scholar] [CrossRef]
- Clementz, M.A.; Chen, Z.; Banach, B.S.; Wang, Y.; Sun, L.; Ratia, K.; Baez-Santos, Y.M.; Wang, J.; Takayama, J.; Ghosh, A.K.; et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010, 84, 4619–4629. [Google Scholar] [CrossRef]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.D.; Baric, R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κ B signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef]
- Zhao, J.; He, S.; Minassian, A.; Li, J.; Feng, P. Recent advances on viral manipulation of NF-κB signaling pathway. Curr. Opin. Virol. 2015, 15, 103–111. [Google Scholar] [CrossRef]
- Rahman, M.M.; McFadden, G. Modulation of NF-κ B signalling by microbial pathogens. Nat. Rev. Microbiol. 2011, 9, 291–306. [Google Scholar] [CrossRef]
- Dong, X.; Feng, H.; Sun, Q.; Li, H.; Wu, T.T.; Sun, R.; Tibbetts, S.A.; Chen, Z.J.; Feng, P. Murine gamma herpesvirus 68 hijacks MAVS and IKKβ to initiate lytic replication. PLoS Pathog. 2010, 6, e1001001. [Google Scholar] [CrossRef]
- Verhoef, K.; Sanders, R.W.; Fontaine, V.; Kitajima, S.; Berkhout, B. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-κ B enhancer element into a GABP binding site. J. Virol. 1999, 73, 1331–1340. [Google Scholar] [CrossRef]
- Wang, J.; Tan, J.; Guo, H.; Zhang, Q.; Jia, R.; Xu, X.; Geng, Y.; Qiao, W. Bovine foamy virus transactivator BTas interacts with cellular RelB to enhance viral transcription. J. Virol. 2010, 84, 11888–11897. [Google Scholar] [CrossRef]
- Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, e1008737. [Google Scholar] [CrossRef]
- Neufeldt, C.J.; Cerikan, B.; Cortese, M.; Frankish, J.; Lee, J.Y.; Plociennikowska, A.; Heigwer, F.; Prasad, V.; Joecks, S.; Burkart, S.S.; et al. SARS-CoV-2 infection induces a pro-infammatory cytokine response through cGAS-STING and NF-κB. BioRxiv 2020. [Google Scholar] [CrossRef]
- Liu, K.; Fang, Y.Y.; Deng, Y.; Liu, W.; Wang, M.F.; Ma, J.P.; Xiao, W.; Wang, Y.N.; Zhong, M.H.; Li, C.H.; et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. 2020, 133, 1025–1031. [Google Scholar] [CrossRef]
- Chen, L.F.; Greene, W.C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 2004, 5, 392–401. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, K.; Wang, D.; Yue, X.; Song, D.; Zhu, Y.; Wu, J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-κB. Virology 2007, 365, 324–335. [Google Scholar] [CrossRef]
- Siu, K.L.; Yuen, K.S.; Castaño-Rodriguez, C.; Ye, Z.W.; Yeung, M.L.; Fung, S.Y.; Yuan, S.; Chan, C.P.; Yuen, K.Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, Z.H.; Lawson, S.R.; Fang, Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 2010, 84, 7832–7846. [Google Scholar] [CrossRef]
- Tofaute, M.J.; Weller, B.; Graß, C.; Halder, H.; Dohai, B.; Falter-Braun, P.; Krappmann, D. SARS-CoV-2 NSP14 MTase activity is critical for inducing canonical NF-κB activation. Biosci. Rep. 2024, 44, BSR20231418. [Google Scholar] [CrossRef]
- Lam, M.T.Y.; Duttke, S.H.; Odish, M.F.; Le, H.D.; Hansen, E.A.; Nguyen, C.T.; Trescott, S.; Kim, R.; Deota, S.; Chang, M.W.; et al. Dynamic activity in cis-regulatory elements of leukocytes identifies transcription factor activation and stratifies COVID-19 severity in ICU patients. Cell Rep. Med. 2023, 4, 100935. [Google Scholar] [CrossRef]
- D’Cruz, L.M.; Knell, J.; Fujimoto, J.K.; Goldrath, A.W. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 2010, 11, 240–249. [Google Scholar] [CrossRef]
- Park, S.H.; Kang, K.; Giannopoulou, E.; Qiao, Y.; Kang, K.; Kim, G.; Park-Min, K.H.; Ivashkiv, L.B. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 2017, 18, 1104–1116. [Google Scholar] [CrossRef]
- Yukawa, M.; Jagannathan, S.; Vallabh, S.; Kartashov, A.V.; Chen, X.; Weirauch, M.T.; Barski, A. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 2020, 217, e20182009. [Google Scholar] [CrossRef]
- Shin, H.M.; Minter, L.M.; Cho, O.H.; Gottipati, S.; Fauq, A.H.; Golde, T.E.; Sonenshein, G.E.; Osborne, B.A. Notch1 augments NF-kB activity by facilitating its nuclear retention. EMBO J. 2006, 25, 129–138. [Google Scholar] [CrossRef]
- Poulsen, L.L.C.; Edelmann, R.J.; Krüger, S.; Diéguez-Hurtado, R.; Shah, A.; Stav-Noraas, T.E.; Renzi, A.; Szymanska, M.; Wang, J.; Ehling, M.; et al. Inhibition of endothelial NOTCH1 signaling attenuates inflammation by reducing cytokine-mediated histone acetylation at inflammatory enhancers. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 854–869. [Google Scholar] [CrossRef]
- Ogawa, S.; Lozach, J.; Benner, C.; Pascual, G.; Tangirala, R.K.; Westin, S.; Hoffmann, A.; Subramaniam, S.; David, M.; Rosenfeld, M.G.; et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 2005, 122, 707–721. [Google Scholar] [CrossRef]
- Kusnadi, A.; Ramírez-Suástegui, C.; Fajardo, V.; Chee, S.J.; Meckiff, B.J.; Simon, H.; Pelosi, E.; Seumois, G.; Ay, F.; Vijayanand, P.; et al. Severely ill patients with COVID-19 display impaired exhaustion features in SARS-CoV-2–reactive CD8+ T cells. Sci. Immunol. 2021, 6, eabe4782. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Whalley, J.P.; Knight, J.C.; Wicker, L.S.; Todd, J.A.; Ferreira, R.C. SARS-CoV-2 infection induces a long-lived proinflammatory transcriptional profile. Genome Med. 2023, 15, 69. [Google Scholar] [CrossRef]
- Gasparello, J.; Marzaro, G.; Papi, C.; Gentili, V.; Rizzo, R.; Zurlo, M.; Scapoli, C.; Finotti, A.; Gambari, R. Effects of Sulforaphane on SARS-CoV-2 infection and NF-κB dependent expression of genes involved in the COVID-19 ‘cytokine storm’. Int. J. Mol. Med. 2023, 52, 76. [Google Scholar] [CrossRef]
- Xu, G.; Qi, F.; Li, H.; Yang, Q.; Wang, H.; Wang, X.; Liu, X.; Zhao, J.; Liao, X.; Liu, Y.; et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020, 6, 73. [Google Scholar] [CrossRef]
- Liu, T.; Jia, P.; Fang, B.; Zhao, Z. Differential Expression of Viral Transcripts from Single-Cell RNA Sequencing of Moderate and Severe COVID-19 Patients and Its Implications for Case Severity. Front. Microbiol. 2020, 11, 603509. [Google Scholar] [CrossRef]
- Li, Z.; Pan, H.; Yang, J.; Chen, D.; Wang, Y.; Zhang, H.; Cheng, Y. Xuanfei Baidu formula alleviates impaired mitochondrial dynamics and activated NLRP3 inflammasome by repressing NF-κB and MAPK pathways in LPS-induced A.L.I. and inflammation models. Phytomedicine 2023, 108, 154545. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ye, L.; Ye, L.; Li, B.; Gao, B.; Zeng, Y.; Kong, L.; Fang, X.; Zheng, H.; Wu, Z.; et al. Up-regulation of IL-6 and TNF-α induced by SARS-coronavirus spike protein in murine macrophages via NF-κB pathway. Virus Res. 2007, 128, 1–8. [Google Scholar] [CrossRef]
- Ma-Lauer, Y.; Carbajo-Lozoya, J.; Hein, M.Y.; Müller, M.A.; Deng, W.; Lei, J.; Meyer, B.; Kusov, Y.; von Brunn, B.; Bairad, D.R.; et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. USA 2016, 113, E5192–E5201. [Google Scholar] [CrossRef]
- Hemmat, N.; Asadzadeh, Z.; Ahangar, N.K.; Alemohammad, H.; Najafzadeh, B.; Derakhshani, A.; Baghbanzadeh, A.; Baghi, H.B.; Javadrashid, D.; Najafi, S.; et al. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch. Virol. 2021, 166, 675–696. [Google Scholar] [CrossRef]
- Smits, S.L.; de Lang, A.; van den Brand, J.M.; Leijten, L.M.; van IJcken, W.F.; Eijkemans, M.J.; van Amerongen, G.; Kuiken, T.; Andeweg, A.C.; Osterhaus, A.D.; et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010, 6, e1000756. [Google Scholar] [CrossRef]
- Hiscott, J.; Nguyen, T.L.; Arguello, M.; Nakhaei, P.; Paz, S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 2006, 25, 6844–6867. [Google Scholar] [CrossRef]
- Clark, J.; Vagenas, P.; Panesar, M.; Cope, A.P. What does tumour necrosis factor excess do to the immune system long term? Ann. Rheum. Dis. 2005, 64 (Suppl. 4), iv70–iv76. [Google Scholar] [CrossRef]
- Silva, L.C.; Ortigosa, L.C.; Benard, G. Anti-TNF-α agents in the treatment of immune-mediated inflammatory diseases: Mechanisms of action and pitfalls. Immunotherapy 2010, 2, 817–833. [Google Scholar] [CrossRef]
- Lapadula, G.; Marchesoni, A.; Armuzzi, A.; Blandizzi, C.; Caporali, R.; Chimenti, S.; Cimaz, R.; Cimino, L.; Gionchetti, P.; Girolomoni, G.; et al. Adalimumab in the treatment of immune-mediated diseases. Int. J. Immunopathol. Pharmacol. 2014, 27 (Suppl. 1), 33–48. [Google Scholar] [CrossRef]
- Dvorak, H.F.; Brown, L.F.; Detmar, M.; Dvorak, A.M. Vascular permeability factor/vascular endothelial growth factor, microvascular hyperpermeability, and angiogenesis. Am. J. Pathol. 1995, 146, 1029. [Google Scholar]
- Charles, P.; Elliott, M.J.; Davis, D.; Potter, A.; Kalden, J.R.; Antoni, C.; Breedveld, F.C.; Smolen, J.S.; Eberl, G.; deWoody, K.; et al. Regulation of cytokines, cytokine inhibitors, and acute-phase proteins following anti-TNF-α therapy in rheumatoid arthritis. J. Immunol. 1999, 163, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Maini, R.N. Anti-TNFα therapy of rheumatoid arthritis: What have we learned? Annu. Rev. Immunol. 2001, 19, 163–196. [Google Scholar] [CrossRef]
- Haga, S.; Yamamoto, N.; Nakai-Murakami, C.; Osawa, Y.; Tokunaga, K.; Sata, T.; Yamamoto, N.; Sasazuki, T.; Ishizaka, Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc. Natl. Acad. Sci. USA 2008, 105, 7809–7814. [Google Scholar] [CrossRef]
- Galloway, J.B.; Hyrich, K.L.; Mercer, L.K.; Dixon, W.G.; Fu, B.; Ustianowski, A.P.; Watson, K.D.; Lunt, M.; Symmons, D.P.; BSRBR Control Centre Consortium; et al. Anti-TNF therapy is associated with an increased risk of serious infections in patients with rheumatoid arthritis especially in the first 6 months of treatment: Updated results from the British Society for Rheumatology Biologics Register with special emphasis on risks in the elderly. Rheumatology 2011, 50, 124–131. [Google Scholar]
- Higuchi, M.; Singh, S.; Chan, H.; Aggarwal, B.B. Aggarwal, Protease inhibitors differentially regulate tumor necrosis factor-induced apoptosis, nuclear factor-κ B activation, cytotoxicity, and differentiation. Blood 1995, 86, 2248–2256. [Google Scholar] [CrossRef]
- D’Acquisto, F.; Sautebin, L.; Iuvone, T.; Di Rosa, M.; Carnuccio, R. Prostaglandins prevent inducible nitric oxide synthase protein expression by inhibiting nuclear factor-κB activation in J774 macrophages. FEBS Lett. 1998, 440, 76–80. [Google Scholar] [CrossRef]
- Rossi, A.; Elia, G.; Santoro, M.G. Activation of the heat shock factor 1 by serine protease inhibitors. An effect associated with nuclear factor-κB inhibition. J. Biol. Chem. 1998, 273, 16446–16452. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta 2010, 1799, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Pancreatic injuries in rats with fecal peritonitis: Protective effect of a new synthetic protease inhibitor, sepinostat mesilate (FUT-187). J. Surg. Res. 1996, 61, 301–306. [Google Scholar] [CrossRef]
- Takahashi, H.; Muto, S.; Nakazawa, E.; Yanagiba, S.; Masunaga, Y.; Miyata, Y.; Tamba, K.; Kusano, E.; Matsuo, M.; Matsuo, T.; et al. Combined treatment with nafamostat mesilate and aspirin prevents heparin-induced thrombocytopenia in a hemodialysis patient. Clin. Nephrol. 2003, 59, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Wang, S.D.; Zeng, Z.Y.; Lin, K.J.; Kao, S.T.; Tani, T.; Yu, C.K.; Wang, J.Y. Serine protease inhibitors nafamostat mesilate and gabexate mesilate attenuate allergen-induced airway inflammation and eosinophilia in a murine model of asthma. J. Allergy Clin. Immunol. 2006, 118, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, M.; Tanaka, H.; Kajiwara, D.; Toyohara, T.; Wakahara, K.; Inagaki, N.; Nagai, H. Nafamostat mesilate, a potent serine protease inhibitor, inhibits airway eosinophilic inflammation and airway epithelial remodeling in a murine model of allergic asthma. J. Pharmacol. Sci. 2008, 108, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Zhirnov, O.P.; Klenk, H.D.; Wright, P.F. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011, 92, 27–36. [Google Scholar] [CrossRef]
- Lin, C.C.; Lin, L.J.; Wang, S.D.; Chiang, C.J.; Chao, Y.P.; Lin, J.; Kao, S.T. The effect of serine protease inhibitors on airway inflammation in a chronic allergen-induced asthma mouse model. Mediators Inflamm. 2014, 2014, 879326. [Google Scholar] [CrossRef]
- Yamaya, M.; Shimotai, Y.; Hatachi, Y.; Kalonji, N.L.; Tando, Y.; Kitajima, Y.; Matsuo, K.; Kubo, H.; Nagatomi, R.; Hongo, S.; et al. The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells. Pulm. Pharmacol. Ther. 2015, 33, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Florencio, A.C.; de Almeida, R.S.; Arantes-Costa, F.M.; Saraiva-Romanholo, B.M.; Duran, A.F.; Sasaki, S.D.; Martins, M.A.; Lopes, F.D.; Tibério, I.F.; Leick, E.A. Effects of the serine protease inhibitor rBmTI-A in an experimental mouse model of chronic allergic pulmonary inflammation. Sci. Rep. 2019, 9, 12624. [Google Scholar] [CrossRef] [PubMed]
- Yuksel, M.; Okajima, K.; Uchiba, M.; Okabe, H. Gabexate mesilate, a synthetic protease inhibitor, inhibits lipopolysaccharide-induced tumor necrosis factor-α production by inhibiting activation of both nuclear factor-κB and activator protein-1 in human monocytes. J. Pharmacol. Exp. Ther. 2003, 305, 298–305. [Google Scholar] [CrossRef]
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 84. [Google Scholar] [CrossRef]
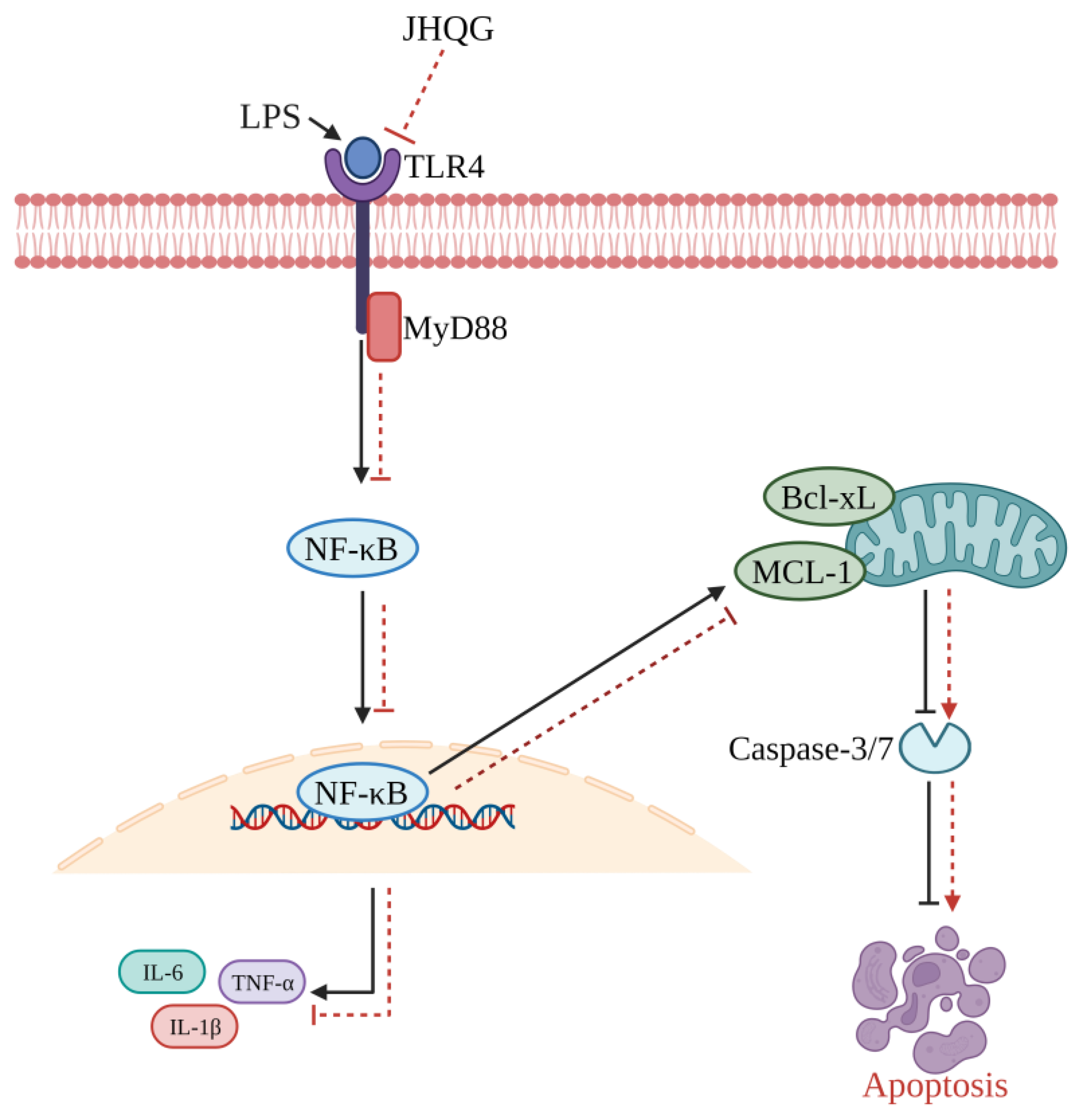
- Zhu, Y.; Han, Q.; Wang, L.; Wang, B.; Chen, J.; Cai, B.; Wu, C.; Zhu, X.; Liu, F.; Han, D.; et al. Jinhua Qinggan granules attenuates acute lung injury by promotion of neutrophil apoptosis and inhibition of TLR4/MyD88/NF-κB pathway. J. Ethnopharmacol. 2023, 301, 115763. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Chen, K.; Gao, Y.; Zhou, X.; Tian, Z.; Chen, W.; Xu, Y.; Chen, Z.; Ning, X. Bioengineered Neutrophil Extinguisher Targets Cascade Immune Pathways of Macrophages for Alleviating Cytokine Storm in Pneumonia. ACS Nano 2023, 17, 16461–16477. [Google Scholar] [CrossRef]
- Biji, A.; Khatun, O.; Swaraj, S.; Narayan, R.; Rajmani, R.S.; Sardar, R.; Satish, D.; Mehta, S.; Bindhu, H.; Jeevan, M.; et al. Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples. EBioMedicine 2021, 70, 103525. [Google Scholar] [CrossRef]
- Rothan, H.A.; Stone, S.; Natekar, J.; Kumari, P.; Arora, K.; Kumar, M. The FDA approved gold drug auranofin inhibits novel coronavirus (SARS-CoV-2) replication and attenuates inflammation in human cells. Virology 2020, 547, 7–11. [Google Scholar] [CrossRef]
- Laplantine, E.; Chable-Bessia, C.; Oudin, A.; Swain, J.; Soria, A.; Merida, P.; Gourdelier, M.; Mestiri, S.; Besseghe, I.; Bremaud, E.; et al. The FDA-approved drug Auranofin has a dual inhibitory effect on SARS-CoV-2 entry and NF-κB signaling. iScience 2022, 25, 105066. [Google Scholar] [CrossRef]
- Jeon, K.I.; Byun, M.S.; Jue, D.M. Gold compound auranofin inhibits IκB kinase (IKK) by modifying Cys-179 of IKKβ subunit. Exp. Mol. Med. 2003, 35, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Saadane, A.; Masters, S.; DiDonato, J.; Li, J.; Berger, M. Parthenolide inhibits IκB kinase, NF-κB activation, and inflammatory response in cystic fibrosis cells and mice. Am. J. Respir. Cell Mol. Biol. 2007, 36, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, P.C.; Behrmann, I.; Müller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334, 297–314. [Google Scholar] [CrossRef]
- Grant, A.H.; Estrada, A., III; Ayala-Marin, Y.M.; Alvidrez-Camacho, A.Y.; Rodriguez, G.; Robles-Escajeda, E.; Cadena-Medina, D.A.; Rodriguez, A.C.; Kirken, R.A. The Many Faces of JAKs and STATs Within the COVID-19 Storm. Front. Immunol. 2021, 12, 690477. [Google Scholar] [CrossRef]
- Gatti, M.; Turrini, E.; Raschi, E.; Sestili, P.; Fimognari, C. Janus Kinase Inhibitors and Coronavirus Disease (COVID)-19: Rationale, Clinical Evidence and Safety Issues. Pharmaceuticals 2021, 14, 738. [Google Scholar] [CrossRef]
- Farahani, M.; Niknam, Z.; Amirabad, L.M.; Amiri-Dashatan, N.; Koushki, M.; Nemati, M.; Pouya, F.D.; Rezaei-Tavirani, M.; Rasmi, Y.; Tayebi, L. Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets. Biomed. Pharmacother. 2022, 145, 112420. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Larcher, A.; Tomelleri, A.; Campochiaro, C.; Della-Torre, E.; De Luca, G.; Farina, N.; Boffini, N.; Ruggeri, A.; Poli, A.; et al. Interleukin-1 and interleukin-6 inhibition compared with standard management in patients with COVID-19 and hyperinflammation: A cohort study. Lancet Rheumatol. 2021, 3, e253–e261. [Google Scholar] [CrossRef] [PubMed]
- Doyle, E.B.; Bentley, D.; Dodds, M.G. COVID-19 acute respiratory distress syndrome: A simulation study of the effects of combination therapy with tocilizumab and siltuximab. Br. J. Clin. Pharmacol. 2022, 88, 1043–1053. [Google Scholar] [CrossRef]
- Pawar, K.S.; Mastud, R.N.; Pawar, S.K.; Pawar, S.S.; Bhoite, R.R.; Bhoite, R.R.; Kulkarni, M.V.; Deshpande, A.R. Oral curcumin with piperine as adjuvant therapy for the treatment of COVID-19: A randomized clinical trial. Front. Pharmacol. 2021, 12, 1056. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kesika, P.; Thangaleela, S.; Sisubalan, N.; Radha, A.; Sivamaruthi, B.S.; Chaiyasut, C. The Role of the Nuclear Factor-Kappa B (NF-κB) Pathway in SARS-CoV-2 Infection. Pathogens 2024, 13, 164. https://doi.org/10.3390/pathogens13020164
Kesika P, Thangaleela S, Sisubalan N, Radha A, Sivamaruthi BS, Chaiyasut C. The Role of the Nuclear Factor-Kappa B (NF-κB) Pathway in SARS-CoV-2 Infection. Pathogens. 2024; 13(2):164. https://doi.org/10.3390/pathogens13020164
Chicago/Turabian StyleKesika, Periyanaina, Subramanian Thangaleela, Natarajan Sisubalan, Arumugam Radha, Bhagavathi Sundaram Sivamaruthi, and Chaiyavat Chaiyasut. 2024. "The Role of the Nuclear Factor-Kappa B (NF-κB) Pathway in SARS-CoV-2 Infection" Pathogens 13, no. 2: 164. https://doi.org/10.3390/pathogens13020164





