Species-Specific Impact of Fusarium Infection on the Root and Shoot Characteristics of Asparagus
Abstract
:1. Introduction
2. Results
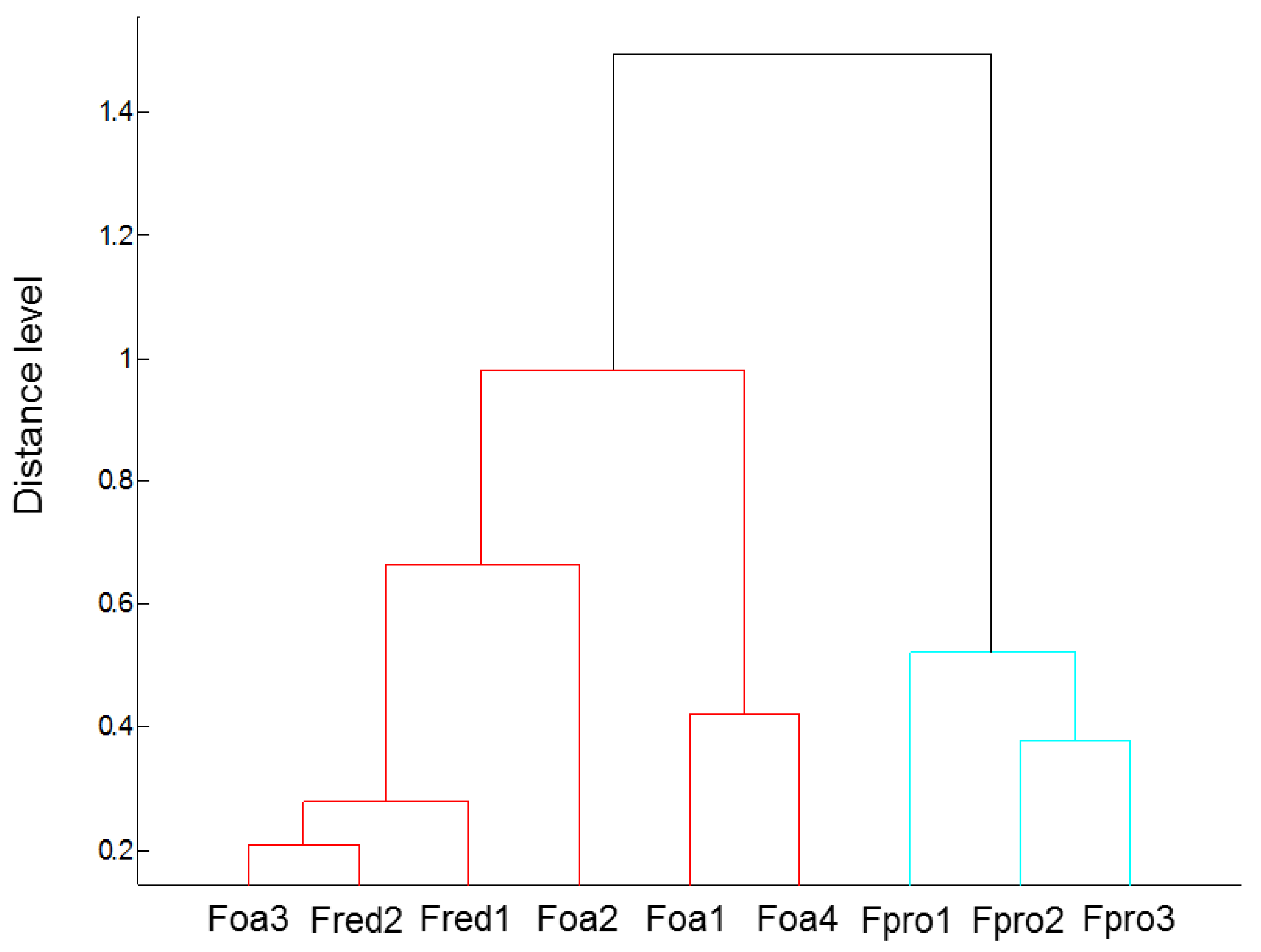
2.1. Identification and Physiological Characterisation of Fusarium Spp.
2.2. Aggressiveness of Fusarium spp. Isolates on Asparagus Plants
2.3. Molecular Detection of Fusarium spp. in Stem Tissue
2.4. Expression of Defence-Related Genes
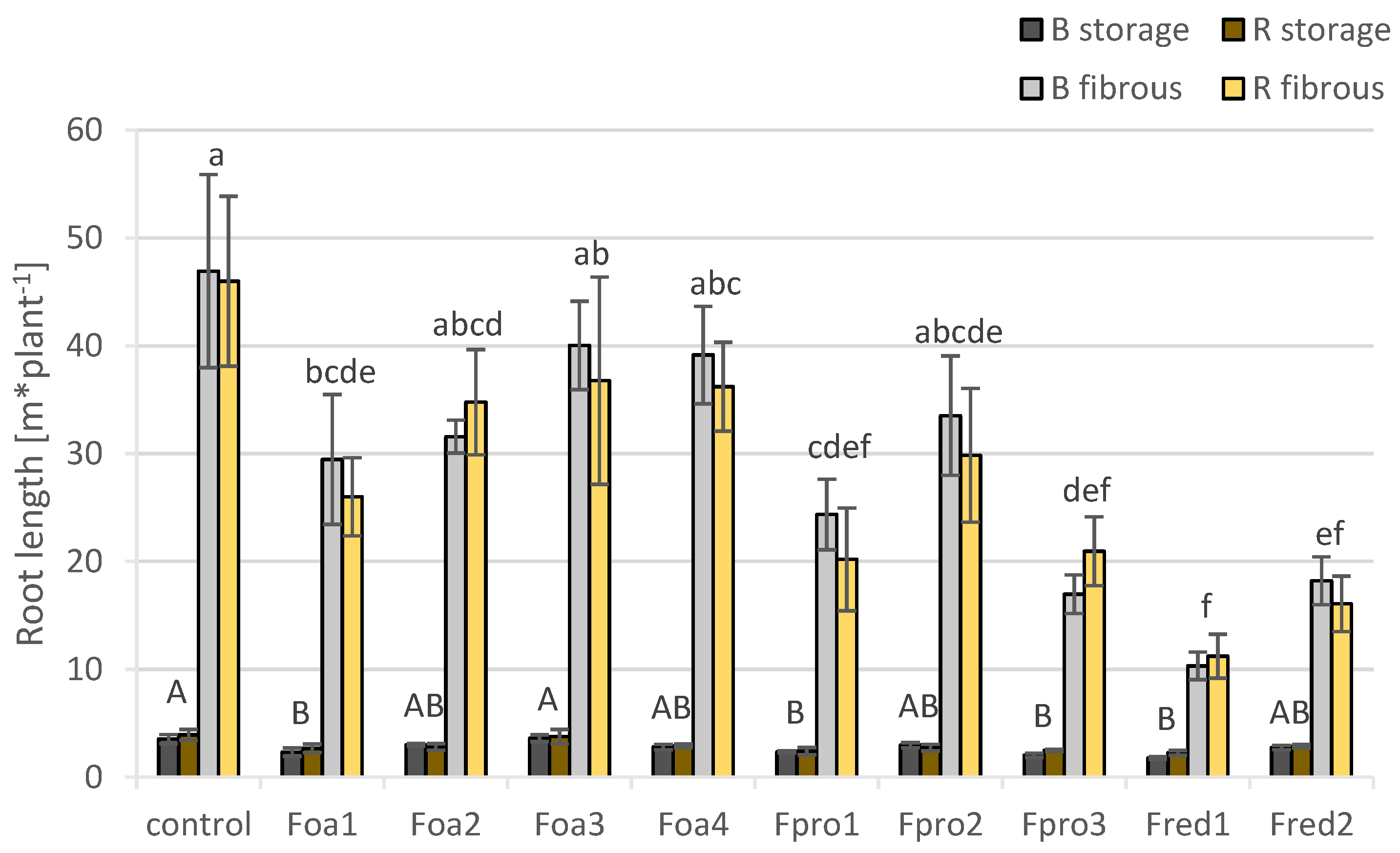
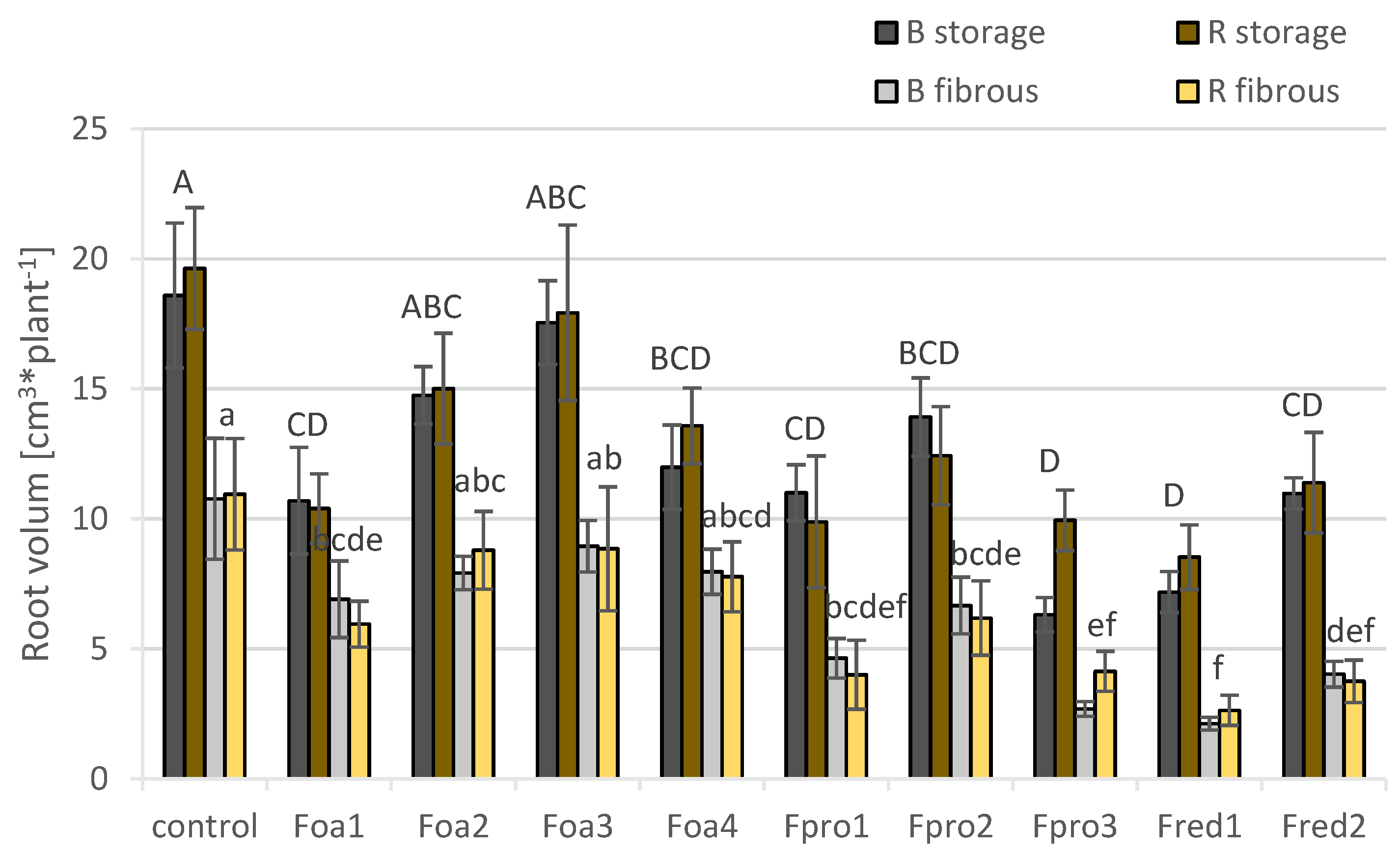
2.5. Changes in Root Morphology
3. Discussion
4. Materials and Methods
4.1. Fusarium spp. Cultivation
4.2. Fungal Degradation of Polysaccharides and Production of Extracellular Hydrolytic Enzymes
4.3. Species-Discrimination by MALDI-TOF Mass Spectrometry
4.4. Plant Cultivation
4.5. Plant Inoculation and Pathogenicity Testing of Fusarium Spp. Isolates on Asparagus Plants
4.6. Molecular Detection of Fusarium Spp. Isolates in Plant Tissue
4.7. Gene Expression Analysis in Stem Tissue Using RT-qPCR
4.8. Analysis of the Root Morphology
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grogan, R.G.; Kimble, K.A. The association of Fusarium wilt with the asparagus decline and replant problem in California. Phytopathology 1959, 49, 122–125. [Google Scholar]
- Elmer, W.H. Combining nonpathogenic strains of Fusarium oxysporum with sodium chloride to suppress Fusarium crown rot of asparagus in replanted fields. Plant Pathol. 2004, 53, 751–758. [Google Scholar] [CrossRef]
- Elmer, W.H. Fusarium diseases of asparagus. In Fusarium: Ecology, Biology & Genetics, Paul E. Nelson Memorial Symposium; Summerell, B.A., Leslie, J.F., Backhouse, D., Bryden, W.L., Burgess, L.W., Eds.; APS Press: St. Paul, MN, USA, 2001; pp. 248–262. [Google Scholar]
- Schreuder, W.; Lamprecht, S.C.; Marasas, W.F.O.; Calitz, F.J. Pathogenicity of three Fusarium species associated with asparagus decline in South Africa. Plant Dis. 1995, 79, 177–181. [Google Scholar] [CrossRef]
- Vujanovic, V.; Hamel, C.; Yergeau, E.; St-Arnaud, M. Biodiversity and biogeography of Fusarium species from northeastern North American asparagus fields based on microbiological and molecular approaches. Microb. Ecol. 2006, 51, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Weber, Z.; Kostecki, M.; von Bargen, S.; Gossmann, M.; Waskiewicz, A.; Bocianowski, J.; Knaflewski, M.; Büttner, C.; Golinski, P. Fusarium Species Colonizing Spears and Forming Mycotoxins in Field Samples of Asparagus from Germany and Poland. J. Phytopathol. 2006, 154, 209–216. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Basallote-Ureba, M.J.; Melero-Vara, J.M.; Abbasi, P.A. Characterization of Fusarium isolates from asparagus fields in southwestern Ontario and influence of soil organic amendments on Fusarium crown and root rot. Phytopathology 2014, 104, 403–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmer, W.H. Management of Fusarium crown and root rot of asparagus. Crop Prot. 2015, 73, 2–6. [Google Scholar] [CrossRef]
- Baayen, R.P.; O′Donnell, K.; Bonants, P.J.; Cigelnik, E.; Kroon, L.P.; Roebroeck, E.J.; Waalwijk, C. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 2000, 90, 891–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fravel, D.; Olivain, C.; Alabouvette, C. Fusarium oxysporum and its biocontrol. New Phytol. 2003, 157, 493–502. [Google Scholar] [CrossRef]
- Blok, W.J.; Bollen, G.J. Fungi on roots and stem bases of asparagus in the Netherlands: Species and pathogenicity. Eur. J. Plant Pathol. 1995, 101, 15–24. [Google Scholar] [CrossRef]
- Marcellan, O.; Camadro, E.; Pontaroli, A.C. Contributions of biotechnology to asparagus breeding in Argentina. Am. J. Plant Sci. Biotech. 2010, 3, 23–30. [Google Scholar]
- Lassaga, S.L.; Camadro, E.L.; Babinec, F.J. Assessing genetic variability for fusarium resistance in three asparagus populations with an in vitro assay. Euphytica 1998, 103, 131–136. [Google Scholar] [CrossRef]
- Damicone, J.P.; Manning, W.J. Avirulent strains of Fusarium oxysporum protect asparagus seedlings from crown rot. Can. J. Plant Pathol. 1982, 4, 143–146. [Google Scholar] [CrossRef]
- He, C.Y.; Hsiang, T.; Wolyn, D.J. Induction of systemic disease resistance and pathogen defence responses in Asparagus officinalis inoculated with nonpathogenic strains of Fusarium oxysporum. Plant Pathol. 2002, 51, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Reid, P.E.; Constable, G.A.; Stiller, W.; Allen, S.; McNamara, G. The CSIRO Fusarium breeding program. In Field to Fashion, Proceedings of the 11th Australian Cotton Conference, Brisbane, 13–15 August 2002; Australian Cotton Growers Research Association: Orange, Australia, 2002; pp. 653–656. [Google Scholar]
- Tu, C.C.; Cheng, Y.H.; Cheng, A.S. Recent advance in biological control of Fusarium wilt of asparagus in Taiwan. Acta Hortic. 1990, 271, 353–362. [Google Scholar] [CrossRef]
- Blok, W.J.; Zwankhuizen, M.J.; Bollen, G.J. Biological control of Fusarium oxysporum f.sp. asparagi by applying non-pathogenic isolates of F. oxysporum. Biocontrol Sci. Technol. 1997, 7, 527–542. [Google Scholar] [CrossRef]
- Arriola, L.L.; Hausbeck, M.K.; Rogers, J.; Safir, G.R. The effect of Trichoderma harzianum and arbuscular mycorrhizae on Fusarium root rot in asparagus. HortTechnology 2000, 10, 141–144. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, Y.; Ohba, N.; Fukui, H. Effect of arbuscular mycorrhizal fungus infection on the incidence of fusarium root rot in asparagus seedlings. J. Jpn. Soc. Hortic. Sci. 2001, 70, 202–206. [Google Scholar] [CrossRef]
- Matsubara, Y.; Hasegawa, N.; Fukui, H. Incidence of Fusarium root rot in asparagus seedlings infected with arbuscular mycorrhizal fungus as affected by several soil amendments. J. Jpn. Soc. Hortic. Sci. 2002, 71, 370–374. [Google Scholar] [CrossRef]
- Wacker, T.L.; Safir, G.R.; Stephens, C.T. Effect of Glomus fasciculatum on the growth of asparagus and the incidence of Fusarium root rot. J. Amer. Soc. Hort. Sci. 1990, 115, 550–554. [Google Scholar] [CrossRef] [Green Version]
- Geiser, D.M.; del Mar Jiménez-Gasco, M.; Kang, S.; Makalowska, I.; Veeraraghavan, N.; Ward, T.J.; Zhang, N.; Kuldau, G.A.; O’Donnell, K. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. In Molecular Diversity and PCR-detection of Toxigenic Fusarium Species and Ochratoxigenic Fungi: Under the aegis of COST Action 835 ‘Agriculturally Important Toxigenic Fungi 1998–2003’, EU project (QLK1-CT-1998-01380) and the ISPP ‘Fusarium Committee’; Mulè, G., Bailey, J.A., Cooke, B.M., Logrieco, A., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 473–479. [Google Scholar]
- Arif, M.; Chawla, S.; Zaidi, N.; Rayar, J.; Variar, M.; Singh, U. Development of specific primers for genus Fusarium and F. solani using rDNA sub-unit and transcription elongation factor (TEF-1α) gene. Afr. J. Biotechnol. 2012, 11, 444–447. [Google Scholar]
- Zarrin, M.; Ganj, F.; Faramarzi, S. Development of a polymerase chain reaction-restriction fragment length polymorphism method for identification of the Fusarium genus using the transcription elongation factor-1α gene. Biomed. Rep. 2016, 5, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Mulè, G.; Susca, A.; Stea, G.; Moretti, A. A species-specific PCR assay based on the calmodulin partial gene for identification of Fusarium verticillioides, F. proliferatum and F. subglutinans. Eur. J. Plant Pathol. 2004, 110, 495–502. [Google Scholar] [CrossRef]
- Mulè, G.; Susca, A.; Stea, G.; Moretti, A. Specific detection of the toxigenic species Fusarium proliferatum and F. oxysporum from asparagus plants using primers based on calmodulin gene sequences. FEMS Microbiol. Lett. 2004, 230, 235–240. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Filion, M.; Vujanovic, V.; St-Arnaud, M. A PCR-denaturing gradient gel electrophoresis approach to assess Fusarium diversity in asparagus. J. Microbiol. Methods 2005, 60, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.Y.; Jeffries, P. Diversity of pathogenic Fusarium populations associated with asparagus roots in decline soils in Spain and the UK. Plant Pathol. 2006, 55, 331–342. [Google Scholar] [CrossRef]
- Corpas-Hervias, C.; Melero-Vara, J.M.; Molinero-Ruiz, M.L.; Zurera-Muñoz, C.; Basallote-Ureba, M.J. Characterization of isolates of Fusarium spp. obtained from asparagus in Spain. Plant Dis. 2006, 90, 1441–1451. [Google Scholar] [CrossRef] [Green Version]
- Rafique, K.; Kang, S.; Aziz-ud-Din; Mahmood, T.; Imran; Ullah, I.; Mahmood, H. First report of Vascular Wilt on Lentil (Lens culinaris Medikus) Caused by Fusarium redolens in Pakistan. Plant Dis. 2020. [Google Scholar] [CrossRef] [Green Version]
- Huisman, O.C. Interrelations of root growth dynamics to epidemiology of root-invading fungi. Annu. Rev. Phytopathol. 1982, 20, 303–327. [Google Scholar] [CrossRef]
- Buhtz, A.; Hohe, A.; Schwarz, D.; Grosch, R. Effects of Verticillium dahliae on tomato root morphology considering plant growth response and defense. Plant Pathol. 2016, 66. [Google Scholar] [CrossRef]
- Morauf, C.; Steinkellner, S. Fusarium oxysporum f. sp. lycopersici and compost affect tomato root morphology. Eur. J. Plant Pathol. 2015, 143, 385–398. [Google Scholar] [CrossRef]
- Wei, J.; Carroll, R.J.; Harden, K.K.; Wu, G. Comparisons of treatment means when factors do not interact in two-factorial studies. Amino Acids 2012, 42, 2031–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patz, S.; Witzel, K.; Scherwinski, A.-C.; Ruppel, S. Culture dependent and independent analysis of potential probiotic bacterial genera and species present in the phyllosphere of raw eaten produce. Int. J. Mol. Sci. 2019, 20, 3661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemptner, J.; Marchetti-Deschmann, M.; Mach, R.; Druzhinina, I.S.; Kubicek, C.P.; Allmaier, G. Evaluation of matrix-assisted laser desorption/ionization (MALDI) preparation techniques for surface characterization of intact fusarium spores by MALDI linear time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Marchetti-Deschmann, M.; Winkler, W.; Dong, H.J.; Lohninger, H.; Kubicek, C.P.; Allmaier, G. Using spores for Fusarium spp. classification by MALDI-based intact cell/spore mass spectrometry. Food Technol. Biotechnol. 2012, 50, 334–342. [Google Scholar]
- Wigmann, É.F.; Behr, J.; Vogel, R.F.; Niessen, L. MALDI-TOF MS fingerprinting for identification and differentiation of species within the Fusarium fujikuroi species complex. Appl. Microbiol. Biotechnol. 2019, 103, 5323–5337. [Google Scholar] [CrossRef]
- Santos, C.; Ventura, J.A.; Lima, N. New insights for diagnosis of pineapple fusariosis by MALDI-TOF MS technique. Curr. Microbiol. 2016, 73, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.T.; Liu, H.Q.; Wang, C.F.; Xu, J.R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 15. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, G.B.; Di Pietro, A.; Roncero, M.I.G. Combined action of the major secreted exo- and endopolygalacturonases is required for full virulence of Fusarium oxysporum. Mol. Plant Pathol. 2016, 17, 339–353. [Google Scholar] [CrossRef]
- Jaroszuk-Ściseł, J.; Kurek, E.; Słomka, A.; Janczarek, M.; Rodzik, B. Activities of cell wall degrading enzymes in autolyzing cultures of three Fusarium culmorum isolates: Growth-promoting, deleterious and pathogenic to rye (Secale cereale). Mycologia 2011, 103, 929–945. [Google Scholar] [CrossRef]
- Jaroszuk-Ściseł, J.; Kurek, E. Hydrolysis of fungal and plant cell walls by enzymatic complexes from cultures of Fusarium isolates with different aggressiveness to rye (Secale cereale). Arch. Microbiol. 2012, 194, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Elmer, W.H.; Stephens, C.T. Classification of Fusarium oxysporum f. sp. asparagi into vegetatively compatible groups. Phytopathology 1989, 79, 88–93. [Google Scholar] [CrossRef]
- Nahiyan, A.S.M.; Boyer, L.R.; Jeffries, P.; Matsubara, Y.-i. PCR-SSCP analysis of Fusarium diversity in asparagus decline in Japan. Eur. J. Plant Pathol. 2011, 130, 197–203. [Google Scholar] [CrossRef]
- Yadeta, K.; Thomma, B. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.W.; Wang, H.W.; Yang, Z.D.; Kong, L.R. Expression comparisons of pathogenesis-related (PR) genes in wheat in response to infection/infestation by fusarium, yellow dwarf virus (YDV) aphid-transmitted and hessian fly. J. Integr. Agric. 2014, 13, 926–936. [Google Scholar] [CrossRef]
- Camm, E.L.; Towers, G.H.N. Phenylalanine ammonia lyase. Phytochemistry 1973, 12, 961–973. [Google Scholar] [CrossRef]
- Çakır, B.; Gül, A.; Yolageldi, L.; Özaktan, H. Response to Fusarium oxysporum f.sp. radicis-lycopersici in tomato roots involves regulation of SA- and ET-responsive gene expressions. Eur. J. Plant Pathol. 2014, 139, 379–391. [Google Scholar] [CrossRef]
- Crosby, K.M. Impact of Monosporascus cannonballus on root growth of diverse melon varieties and their F1 progeny inthe field. Subtrop. Plant Sci. 2000, 52, 8–11. [Google Scholar]
- Cichy, K.A.; Snapp, S.S.; Kirk, W.W. Fusarium root rot incidence and root system architecture in grafted common bean lines. Plant Soil 2007, 300, 233–244. [Google Scholar] [CrossRef]
- Kraft, J.M.; Boge, W. Root characteristics in pea in relation to compaction and Fusarium root rot. Plant Dis. 2001, 85, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.M.D.; Leandro, L.F.; Munkvold, G.P. Aggressiveness of Fusarium species and impact of root infection on growth and yield of soybeans. Phytopathology 2013, 103, 822–832. [Google Scholar] [CrossRef] [Green Version]
- Elmer, W. Asparagus decline and replant problem: A look back and a look forward at strategies for mitigating losses. Acta Hortic. 2017, 1223, 195–204. [Google Scholar] [CrossRef]
- Guo, J.; Jermyn, W.A.; Turnbull, M.H. Diurnal and seasonal photosynthesis in two asparagus cultivars with contrasting yield. Crop Sci. 2002, 42, 399–405. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Irzykowska, L.; Karolewski, Z.; Bocianowski, J.; Goliński, P.; Weber, Z. Mycotoxins biosynthesis by Fusarium oxysporum and F. proliferatum isolates of asparagus origin. J. Plant Prot. Res. 2009, 49, 3699. [Google Scholar] [CrossRef]
- Shi, W.; Tan, Y.; Wang, S.; Gardiner, D.M.; De Saeger, S.; Liao, Y.; Wang, C.; Fan, Y.; Wang, Z.; Wu, A. Mycotoxigenic potentials of Fusarium species in various culture matrices revealed by mycotoxin profiling. Toxins 2016, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Knaflewski, M.; Golinski, P.; Kostecki, M.; Waskiewicz, A.; Weber, Z. Mycotoxins and mycotoxin-producing fungi occurring in asparagus spears. Acta Horticulturae 2008, 776, 183–191. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Irzykowska, L.; Bocianowski, J.; Karolewski, Z.; Kostecki, M.; Weber, Z.; Goliński, P. Occurrence of Fusarium fungi and mycotoxins in marketable asparagus spears. Polish J. Environ. Stud. 2010, 19, 219–225. [Google Scholar]
- Ismaiel, A.A.; Papenbrock, J. Mycotoxins: Producing fungi and mechanisms of phytotoxicity. Agriculture 2015, 5, 492–537. [Google Scholar] [CrossRef] [Green Version]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, L.; Sánchez-Juanes, F.; Muñoz-Bellido, J.L.; González-Buitrago, J.M. Rapid method for direct identification of bacteria in urine and blood culture samples by matrix-assisted laser desorption ionization time-of-flight mass spectrometry: Intact cell vs. extraction method. Clin. Microbiol. Infect. 2011, 17, 1007–1012. [Google Scholar] [CrossRef] [Green Version]
- Feller, C.; Richter, E.; Smolders, T.; Wichura, A. Phenological growth stages of edible asparagus (Asparagus officinalis): Codification and description according to the BBCH scale. Ann. Appl. Biol. 2012, 160, 174–180. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. (Eds.) The Fusarium Laboratory Manual; Blackwell Publishing Professional: Ames, IA, USA, 2006. [Google Scholar]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierret, A.; Gonkhamdee, S.; Jourdan, C.; Maeght, J.-L. IJ_Rhizo: An open-source software to measure scanned images of root samples. Plant Soil 2013, 373, 531–539. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Brunner, E.; Munzel, U. Nichtparametrische Datenanalyse; Springer Spektrum Berlin: Berlin, Germany, 2013. [Google Scholar]
- Holm, S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]





| Isolate | Origin Code | Species | Origin | Isolation | Source * |
|---|---|---|---|---|---|
| Foa1 | Ob2-66 | F. oxysporum | Brandenburg | 2002 | HUB |
| Foa2 | 10K3.3 | F. oxysporum | Niedersachsen | 2001 | HUB |
| Foa3 | 45K/2.1 | F. oxysporum | Niedersachsen | 2001 | HUB |
| Foa4 | III/3.1 | F. oxysporum | Sachsen-Anhalt | 2011 | JKI |
| Fpro1 | 193-S | F. proliferatum | Brandenburg | 2002 | HUB |
| Fpro2 | 219-S | F. proliferatum | Rheinland-Pfalz | 2000 | HUB |
| Fpro3 | 88-17/1 | F. proliferatum | Niedersachsen | 2015 | UAS |
| Fred1 | 89-17/2 | F. redolens | Niedersachsen | 2015 | UAS |
| Fred2 | 90-7/1 | F. redolens | Niedersachsen | 2015 | UAS |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djalali Farahani-Kofoet, R.; Witzel, K.; Graefe, J.; Grosch, R.; Zrenner, R. Species-Specific Impact of Fusarium Infection on the Root and Shoot Characteristics of Asparagus. Pathogens 2020, 9, 509. https://doi.org/10.3390/pathogens9060509
Djalali Farahani-Kofoet R, Witzel K, Graefe J, Grosch R, Zrenner R. Species-Specific Impact of Fusarium Infection on the Root and Shoot Characteristics of Asparagus. Pathogens. 2020; 9(6):509. https://doi.org/10.3390/pathogens9060509
Chicago/Turabian StyleDjalali Farahani-Kofoet, Roxana, Katja Witzel, Jan Graefe, Rita Grosch, and Rita Zrenner. 2020. "Species-Specific Impact of Fusarium Infection on the Root and Shoot Characteristics of Asparagus" Pathogens 9, no. 6: 509. https://doi.org/10.3390/pathogens9060509





