Antimicrobial Resistance Surveillance of Tigecycline-Resistant Strains Isolated from Herbivores in Northwest China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolates
2.2. Antimicrobial Susceptibility
2.3. Whole-Genome Sequencing
2.4. Identification of Antimicrobial Resistant Genes, Multilocus Sequence Typing Analysis, and Virulence Factors
2.5. Statistical Analysis
3. Results
3.1. Tigecycline Resistant Isolates
3.2. Antibiotic Sensitivity Test
3.3. Whole Genome Sequencing Analysis
3.4. Distribution of Antimicrobial Resistance Genes and Virulence Factors of Isolates
3.5. Multilocus Sequence Typing (MLST)
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhong, C.; Liu, T.; Zhu, Y.; Gou, S.; Bao, H.; Yao, J.; Ni, J. Newly designed antimicrobial peptides with potent bioactivity and enhanced cell selectivity prevent and reverse rifampin resistance in Gram-negative bacteria. Eur. J. Pharm. Sci. 2021, 158, 105665. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, K.; Fisher, J.F. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 2011, 65, 455–478. [Google Scholar] [CrossRef]
- Tangden, T.; Giske, C.G. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: Clinical perspectives on detection, treatment and infection control. J. Intern. Med. 2015, 277, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.L.; Marks, S.J.; Montealegre, M.C.; Gilman, R.H.; Pajuelo, M.J.; Saito, M.; Tsukayama, P.; Njenga, S.M.; Kiiru, J.; Swarthout, J.; et al. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 2020, 5, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Haworth-Brockman, M.; Saxinger, L.M.; Miazga-Rodriguez, M.; Wierzbowski, A.; Otto, S.J.G. One Health Evaluation of Antimicrobial Use and Resistance Surveillance: A Novel Tool for Evaluating Integrated, One Health Antimicrobial Resistance and Antimicrobial Use Surveillance Programs. Front. Public Health 2021, 9, 693703. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Wang, J.; Zheng, X.; Chang, J.; Ma, J.; Wang, J.; Ji, X.; Yang, H.; Ding, B. Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai Plateau of China. Front. Microbiol. 2022, 13, 885132. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [Green Version]
- Shafiq, M.; Rahman, S.U.; Bilal, H.; Ullah, A.; Noman, S.M.; Zeng, M.; Yuan, Y.; Xie, Q.; Li, X.; Jiao, X. Incidence and molecular characterization of ESBL-producing and colistin-resistant Escherichia coli isolates recovered from healthy food-producing animals in Pakistan. J. Appl. Microbiol. 2022, 133, 1169–1182. [Google Scholar] [CrossRef]
- Li, Y.; Peng, K.; Yin, Y.; Sun, X.; Zhang, W.; Li, R.; Wang, Z. Occurrence and Molecular Characterization of Abundant tet(X) Variants among Diverse Bacterial Species of Chicken Origin in Jiangsu, China. Front. Microbiol. 2021, 12, 751006. [Google Scholar] [CrossRef]
- Zhu, D.K.; Luo, H.Y.; Liu, M.F.; Zhao, X.X.; Jia, R.Y.; Chen, S.; Sun, K.F.; Yang, Q.; Wu, Y.; Chen, X.Y.; et al. Various Profiles of tet Genes Addition to tet(X) in Riemerella anatipestifer Isolates from Ducks in China. Front. Microbiol. 2018, 9, 585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leski, T.A.; Bangura, U.; Jimmy, D.H.; Ansumana, R.; Lizewski, S.E.; Stenger, D.A.; Taitt, C.R.; Vora, G.J. Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int. J. Antimicrob. Agents 2013, 42, 83–86. [Google Scholar] [CrossRef]
- Liu, G.; Qin, M. Analysis of the Distribution and Antibiotic Resistance of Pathogens Causing Infections in Hospitals from 2017 to 2019. Evid. Based Complement. Alternat. Med. 2022, 2022, 3512582. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Dudeja, M.; Kohli, S.; Ray, P. Genotypic characterization of vancomycin-resistant Enterococcus causing urinary tract infection in northern India. Indian J. Med. Res. 2022, 155, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Gordon, N.C.; Wareham, D.W. A review of clinical and microbiological outcomes following treatment of infections involving multidrug-resistant Acinetobacter baumannii with tigecycline. J. Antimicrob. Chemother. 2009, 63, 775–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, H.; Mei, C.Y.; Wang, Y.; Wang, Z.Y.; Lu, M.J.; Pan, Z.M.; Jiao, X. Multiple Mechanisms of Tigecycline Resistance in Enterobacteriaceae from a Pig Farm, China. Microbiol. Spectr. 2021, 9, e0041621. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, H.; Li, Q.; Peng, Q.; Zhao, Q.; Wang, J.; Liu, X. Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolated from the rivers and lakes in Northwest China. BMC Microbiol. 2018, 18, 125. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liu, H.; Wang, L.; Peng, Q.; Li, Y.; Zhou, H.; Li, Q. Molecular Characterization of Extended-Spectrum beta-Lactamase-Producing Multidrug Resistant Escherichia coli from Swine in Northwest China. Front. Microbiol. 2018, 9, 1756. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Shang, Y.; Liu, Y.; Li, Z.; Jing, Z. Genetic Characterization of Animal Brucella Isolates from Northwest Region in China. Biomed. Res. Int. 2018, 2018, 2186027. [Google Scholar] [CrossRef] [Green Version]
- Jo, J.; Ko, K.S. Tigecycline Heteroresistance and Resistance Mechanism in Clinical Isolates of Acinetobacter baumannii. Microbiol. Spectr. 2021, 9, e0101021. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Chang, J.; Zhang, L.; Liu, L.; Xia, X.; Hassan, B.H.; Jia, X.; Yang, H.; Feng, Y. Carriage of Distinct mcr-1-Harboring Plasmids by Unusual Serotypes of Salmonella. Adv. Biosyst. 2020, 4, e1900219. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Zhang, W.; Tang, X.; Xin, Y.; Xu, Y.; Sun, H.; Luo, X.; Pu, J.; Xu, J.; Xiong, Y.; et al. Shiga Toxin-Producing Escherichia coli in Plateau Pika (Ochotona curzoniae) on the Qinghai-Tibetan Plateau, China. Front. Microbiol. 2016, 7, 375. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Tang, B.; Chen, Y.; Xia, X.; Qian, M.; Yang, H. Two IncHI2 Plasmid-Mediated Colistin-Resistant Escherichia coli Strains from the Broiler Chicken Supply Chain in Zhejiang Province, China. J. Food Prot. 2020, 83, 1402–1410. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 2020, 11, e02930-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawalek, A.; Modrzejewska, M.; Zieniuk, B.; Bartosik, A.A.; Jagura-Burdzy, G. Interaction of ArmZ with the DNA-binding domain of MexZ induces expression of mexXY multidrug efflux pump genes and antimicrobial resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, 12. [Google Scholar] [CrossRef] [PubMed]
- Grau, F.C.; Jaeger, J.; Groher, F.; Suess, B.; Muller, Y.A. The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex. Nucleic Acids Res. 2020, 48, 3366–3378. [Google Scholar] [CrossRef]
- Moller, T.S.B.; Overgaard, M.; Nielsen, S.S.; Bortolaia, V.; Sommer, M.O.A.; Guardabassi, L.; Olsen, J.E. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. Bmc Microbiol. 2016, 16, 1–8. [Google Scholar] [CrossRef]
- Nguyen, K.V.; Nguyen, T.V.; Nguyen, H.T.T.; Le, D.V. Mutations in the gyrA, parC, and mexR genes provide functional insights into the fluoroquinolone-resistant Pseudomonas aeruginosa isolated in Vietnam. Infect. Drug Resist. 2018, 11, 275–282. [Google Scholar] [CrossRef] [Green Version]
- El-Badawy, M.F.; Eed, E.M.; Sleem, A.S.; El-Sheikh, A.A.; Maghrabi, I.A.; Abdelwahab, S.F. The First Saudi Report of Novel and Common Mutations in the gyrA and parC Genes among Pseudomonas Spp. Clinical Isolates Recovered from Taif Area. Infect. Drug Resist. 2022, 15, 3801–3814. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, K.; Tanimoto, K.; Fueki, S.; Tomita, H.; Hirakawa, H. Elevated Expression of GlpT and UhpT via FNR Activation Contributes to Increased Fosfomycin Susceptibility in Escherichia coli under Anaerobic Conditions. Antimicrob. Agents Chemother. 2015, 59, 6352–6360. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Fu, Z.Y.J.; Zhou, Y.; Liu, Y.; Xu, X.G.; Wang, M.G. Mutations of the Transporter Proteins GlpT and UhpT Confer Fosfomycin Resistance in Staphylococcus aureus. Front. Microbiol. 2017, 8, 914. [Google Scholar] [CrossRef] [Green Version]
- Patino-Navarrete, R.; Rosinski-Chupin, I.; Cabanel, N.; Gauthier, L.; Takissian, J.; Madec, J.Y.; Hamze, M.; Bonnin, R.A.; Naas, T.; Glaser, P. Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 2020, 12, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domain, F.; Levy, S.B. GyrA Interacts with MarR To Reduce Repression of the marRAB Operon in Escherichia coli. J. Bacteriol. 2011, 193, 2674, Retraction of J. Bacteriol. 2010, 192, 942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warner, D.M.; Yang, Q.W.; Duval, V.; Chen, M.J.; Xu, Y.C.; Levy, S.B. Involvement of MarR and YedS in Carbapenem Resistance in a Clinical Isolate of Escherichia coli from China. Antimicrob. Agents Chemother. 2013, 57, 1935–1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudu, P.; Moon, N.; Weiss, B. Regulation of the soxRS oxidative stress regulon—Reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem. 1997, 272, 5082–5086. [Google Scholar] [CrossRef] [Green Version]
- Koutsolioutsou, A.; Pena-Llopis, S.; Demple, B. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 2005, 49, 2746–2752. [Google Scholar] [CrossRef] [Green Version]
- Subhadra, B.; Kim, J.; Kim, D.H.; Woo, K.; Oh, M.H.; Choi, C.H. Local Repressor AcrR Regulates AcrAB Efflux Pump Required for Biofilm Formation and Virulence in Acinetobacter nosocomialis. Front. Cell. Infect. Microbiol. 2018, 8, 270. [Google Scholar] [CrossRef]
- Jahandideh, S. Diversity in Structural Consequences of MexZ Mutations in Pseudomonas aeruginosa. Chem. Biol. Drug Des. 2013, 81, 600–606. [Google Scholar] [CrossRef] [PubMed]
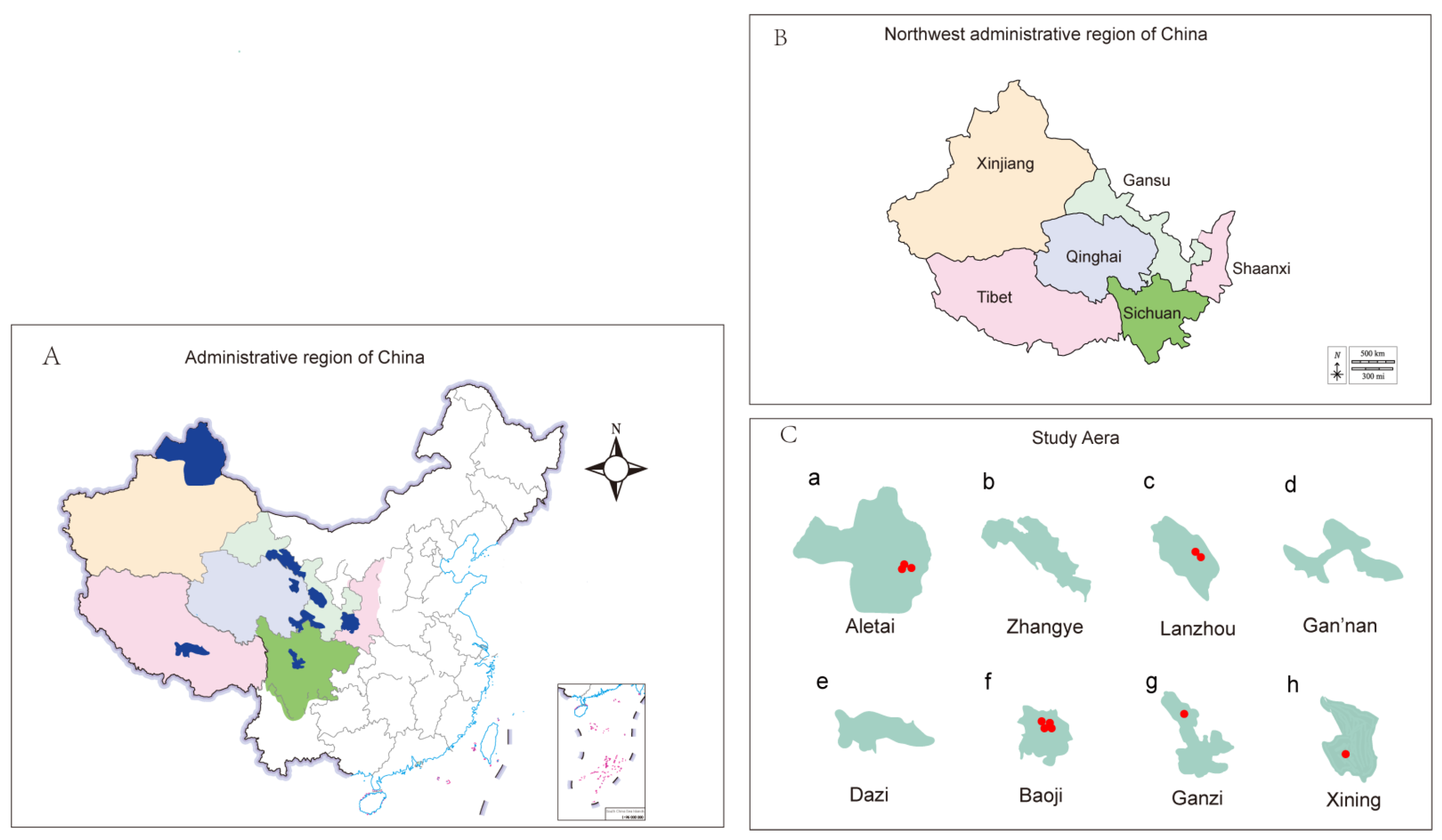


| Region | Study Area | Isolates, n 2021 March–June | 2021 September–December | 2022 March–June | 2022 Seprember–December | Total | Prevalence |
|---|---|---|---|---|---|---|---|
| Xinjiang | Aletai | 60 | - | - | - | 60 | 3/60, 5.0% |
| Gansu | Zhangye | 30 | - | - | - | 30 | 0 |
| Lanzhou | - | - | - | 30 | 30 | 2/30, 6.67% | |
| Gan’nan | - | - | 30 | - | 30 | 0 | |
| Shaanxi | Baoji | - | 30 | 30 | - | 60 | 4/60, 6.67% |
| Sichuan | Ganzi | - | - | - | 30 | 30 | 1/30, 3.33% |
| Tibet | Dazi | 30 | - | - | - | 30 | 0 |
| Qinghai | Xining | - | 30 | - | - | 30 | 1/30, 3.33% |
| Antimicrobial Agents | Bacterial Isolates (MIC µg/mL) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SX-3E-11 | SX-2E-03 | SX-1E-06 | SX-1E-08 | SC-1E-03 | XJ-2E-05 | XJ-1E-02 | LZ-1S-01 | LZ-1P-09 | QH-2K-03 | XJ-1K-13 | |
| Ampicillin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Amoxicillin/clavulanate potassium | 32 | 32 | 32 | 16 | 32 | 32 | 16 | 16 | 16 | 2 | 4 |
| Gentamicin | 0.5 | 2 | 0.5 | 2 | 8 | 4 | 32 | 4 | 32 | 1 | 8 |
| Cefotaxime | 1 | 4 | 2 | 16 | 32 | 32 | 16 | >256 | 32 | 4 | 2 |
| Meropenem | 0.25 | 0.25 | 0.125 | 0.125 | 0.5 | 1 | 1 | 1 | 0.5 | 0.25 | 0.125 |
| Amikacin | 16 | 32 | 32 | 16 | 32 | 32 | 16 | 128 | 32 | 64 | 8 |
| Ceftiofur | 2 | 0.5 | 1 | 4 | 0.25 | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | 0.25 |
| Colistin | 0.5 | 0.125 | 0.25 | 0.25 | 0.125 | 0.125 | 0.125 | 0.125 | 0.5 | 0.125 | 0.125 |
| Ciprofloxacin | 8 | 0.125 | 0.0625 | 8 | 0.125 | 0.0625 | 8 | 0.0625 | 8 | 0.125 | 8 |
| Sulfamethoxazole | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Tetracycline | 16 | 64 | 64 | 32 | 8 | 16 | 64 | 64 | 64 | 8 | 64 |
| Tigecycline | 8 | 4 | 8 | 8 | 8 | 4 | 8 | 8 | 8 | 32 | 32 |
| Florfenicol | 0.25 | 64 | 64 | 64 | 32 | 8 | 8 | 64 | 8 | 64 | 64 |
| Fosfomycin | 16 | 16 | 16 | 2 | 8 | 16 | 8 | 16 | 2 | 8 | 8 |
| Genomic Data | SX-3E-11 | SX-2E-03 | SX-1E-06 | SX-1E-08 | QH-2K-03 | SC-1E-03 | XJ-2E-05 | XJ-1K-13 | XJ-1E-02 | LZ-1P-09 | LZ-1S-01 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Raw_data | 3,464,897,100 | 3,668,641,500 | 2,981,649,300 | 3,394,535,100 | 3,267,123,600 | 3,229,710,600 | 2,799,241,800 | 3,049,228,200 | 3,552,870,300 | 1,913,231,700 | 3,959,209,500 |
| Raw reeds | 23,099,314 | 24,457,610 | 19,877,662 | 22,630,234 | 21,780,824 | 21,531,404 | 18,661,612 | 20,328,188 | 23,685,802 | 12,754,878 | 26,394,730 |
| Read_length | 150 | 150 | 150 | 150 | 150 | 150 | 150 | 150 | 150 | 150 | 150 |
| Sequence length | 5,154,027 | 5,417,711 | 5,417,297 | 5,071,184 | 5,574,832 | 5,105,116 | 5,066,943 | 6,028,924 | 4,819,548 | 31,174,726 | 4,754,898 |
| Scaffolds count | 127 | 155 | 152 | 104 | 81 | 187 | 129 | 82 | 188 | 364 | 31 |
| % GC a | 50.98 | 50.63 | 50.38 | 50.54 | 56.98 | 50.22 | 50.53 | 55.05 | 51.45 | 56.71 | 52 |
| Q20(%) b | 98.01 | 97.99 | 97.98 | 97.95 | 97.9 | 97.96 | 97.8 | 98.04 | 97.8 | 97.73 | 97.89 |
| Largest contig | 626,493 | 387,573 | 387,573 | 593,011 | 905,104 | 402,856 | 556,679 | 465,690 | 231,092 | 556,019 | 773,785 |
| N50 c | 181,274 | 123,205 | 123,205 | 143,900 | 234,490 | 111,759 | 177,264 | 221,920 | 96,022 | 159,433 | 467,849 |
| N90 d | 32,857 | 30,785 | 31,761 | 39,038 | 54,273 | 25,327 | 34,748 | 70,423 | 18,581 | 57,075 | 152,566 |
| L50 e | 9 | 14 | 14 | 11 | 7 | 14 | 9 | 9 | 17 | 61 | 4 |
| L75 f | 17 | 27 | 27 | 22 | 15 | 30 | 20 | 17 | 34 | 128 | 7 |
| Isolate Name | Allele adK | fumC | gyrB | icd | mdh | purA | recA | ST a | CC b |
|---|---|---|---|---|---|---|---|---|---|
| SX-3E-11 | 6 | 6 | 15 | 56 | 11 | 26 | 6 | 2144 | - |
| SX-2E-03 | 43 | 41 | 15 | 18 | 11 | 7 | 6 | 101 | ST101 Cplx |
| SX-1E-06 | 43 | 41 | 15 | 18 | 11 | 7 | 6 | 101 | ST101 Cplx |
| SX-1E-08 | 10 | 11 | 4 | 8 | 179 | 8 | 2 | 3944 | - |
| SC-1E-03 | 6 | 4 | 1323 | 1 | 9 | 2 | 7 | 13667 | - |
| XJ-2E-05 | 6 | 4 | 4 | 16 | 24 | 8 | 14 | 58 | ST155 Cplx |
| XJ-1E-02 | 6 | 107 | 1 | 95 | 69 | 8 | 20 | 536 | ST399 Cplx |
| gapA | infB | mdh | pgi | phoE | rpoB | tonB | ST | ||
| QH-2K-03 | 2 | 1 | 2 | 1 | 10 | 1 | 12 | 999 | - |
| XJ-1K-13 | 1 | 7 | 2 | 30 | 4 | 1 | 2 | 65 | - |
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | ST | ||
| LZ-1P-09 | 24 | 16 | 20 | 16 | 15 | 14 | 21 | 68 | - |
| aroC | dnaN | hemD | hisD | purE | sucA | thrA | ST | ||
| LZ-1S-01 | 5 | 2 | 3 | 7 | 31 | 41 | 11 | 92 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Shao, C.; Gong, X.; Quan, H.; Liu, D.; Chen, Q.; Chu, Y. Antimicrobial Resistance Surveillance of Tigecycline-Resistant Strains Isolated from Herbivores in Northwest China. Microorganisms 2022, 10, 2432. https://doi.org/10.3390/microorganisms10122432
Yu Y, Shao C, Gong X, Quan H, Liu D, Chen Q, Chu Y. Antimicrobial Resistance Surveillance of Tigecycline-Resistant Strains Isolated from Herbivores in Northwest China. Microorganisms. 2022; 10(12):2432. https://doi.org/10.3390/microorganisms10122432
Chicago/Turabian StyleYu, Yongfeng, Changchun Shao, Xiaowei Gong, Heng Quan, Donghui Liu, Qiwei Chen, and Yuefeng Chu. 2022. "Antimicrobial Resistance Surveillance of Tigecycline-Resistant Strains Isolated from Herbivores in Northwest China" Microorganisms 10, no. 12: 2432. https://doi.org/10.3390/microorganisms10122432





