West-Siberian Chernozem: How Vegetation and Tillage Shape Its Bacteriobiome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Conditions
2.2. Experimental Setup
2.3. Soil Sampling and Chemical Analyses
2.4. DNA Extraction, Amplification and Sequencing
2.5. Bioinformatic Analysis
2.6. Statistical Analyses
3. Results
3.1. General Taxonomic Diversity
3.2. Bacterial Taxonomic Diversity as Related to the Experimental Fields
3.3. Bacteriobiome α- and β-Biodiversity
4. Discussion
4.1. Soil Bacteriobiome: General Outline
4.2. The Effect of Soil Tillage on Soil Bacteriobiome
4.3. Bacterial Genera and OTUs That Differentially Increased in the Undisturbed Soil
4.4. General Comments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brussaard, L.; De Ruiter, P.C.; Brown, G.G. Soil biodiversity for agricultural sustainability. Agric. Ecosyst. Environ. 2007, 121, 233–244. [Google Scholar] [CrossRef]
- Lin, J.S.; Sarto, M.V.M.; Carter, T.L.; Peterson, D.E.; Gura, C.; Mino, L.; Rohrs, M.; Lucas, H.; Clark, J.; Rice, C.W. Soil organic carbon, aggregation and fungi community after 44 years of no-till and cropping systems in the Central Great Plains, USA. Arch. Microbiol. 2023, 205, 84. [Google Scholar] [CrossRef] [PubMed]
- Derpsch, R.; Friedrich, T.; Kassam, A.; Li, H. Current status of adoption of no-till farming in the world and some of its main benefits. Int. J. Agric. Biol. Engin. 2010, 3, 1–25. [Google Scholar]
- Abbas, F.; Hammad, H.M.; Ishaq, W.; Farooque, A.A.; Bakhat, H.F.; Zia, Z.; Fahad, S.; Farhad, W.; Cerdà, A. A review of soil carbon dynamics resulting from agricultural practices. J. Environ. Manag. 2020, 268, 110319. [Google Scholar] [CrossRef] [PubMed]
- Behnke, G.D.; Zabaloy, M.C.; Riggins, C.W.; Rodríguez-Zas, S.; Huang, L.; Villamil, M.B. Acidification in corn monocultures favor fungi, ammonia oxidizing bacteria, and nirK-denitrifier groups. Sci. Total Environ. 2020, 720, 137514. [Google Scholar] [CrossRef]
- Cornell, C.R.; Zhang, Y.; Van Nostrand, J.D.; Wagle, P.; Xiao, X.; Zhou, J. Temporal Changes of Virus-Like Particle Abundance and Metagenomic Comparison of Viral Communities in Cropland and Prairie Soils. mSphere 2021, 6, e0116020. [Google Scholar] [CrossRef]
- Legrand, F.; Picot, A.; Cobo-Díaz, J.F.; Carof, M.; Chen, W.; Le Floch, G. Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 2018, 132, 135–145. [Google Scholar] [CrossRef]
- Tyler, H.L. Bacterial community composition under long-term reduced tillage and no till management. J. Appl. Microbiol. 2019, 126, 1797–1807. [Google Scholar] [CrossRef]
- Degrune, F.; Theodorakopoulos, N.; Dufrêne, M.; Colinet, G.; Bodson, B.; Hiel, M.P.; Taminiau, B.; Nezer, C.; Daube, G.; Vandenbol, M. No favorable effect of reduced tillage on microbial community diversity in a silty loam soil (Belgium). Agric. Ecosyst. Environ. 2016, 224, 12–21. [Google Scholar] [CrossRef]
- Naumova, N.; Barsukov, P.; Baturina, O.; Rusalimova, O.; Kabilov, M. Soil Mycobiome Diversity under Different Tillage Practices in the South of West Siberia. Life 2022, 12, 1169. [Google Scholar] [CrossRef]
- IUSS Working Group. WRB, World Reference Base for Soil Resources 2015. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; FAO: Rome, Italy, 2015. [Google Scholar]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Naumova, N.; Alikina, T.; Tupikin, A.; Kalmykova, A.; Soldatova, G.; Vlassov, V.; Kabilov, M. Human Gut Microbiome Response to Short-Term Bifidobacterium-Based Probiotic Treatment. Indian J. Microbiol. 2020, 60, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Fauth, E.; Bernardo, J.; Camara, M.; Resetarits, W.J., Jr.; Van Buskirk, J.; McCollum, S.A. Simplifying the Jargon of Community Ecology: A Conceptual Approach. Am. Nat. 1996, 147, 282–286. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Hughes, J.B.; Hellmann, J.J. The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. Methods Enzymol. 2005, 397, 292–308. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Bendia, A.G.; Fricker, A.D.; Pellizari, V.H.; Galante, D.; Rodrigues, F. Isolation of Uncultured Bacteria from Antarctica Using Long Incubation Periods and Low Nutritional Media. Front. Microbiol. 2017, 8, 1346. [Google Scholar] [CrossRef]
- Vasconcellos, R.L.F.; Romagnoli, E.M.; Taketani, R.G.; Santos, S.N.; Zucchi, T.D.; Melo, I.S. Impact of Inoculation with Pseudomonas aestus CMAA 1215T on the Non-target Resident Bacterial Community in a Saline Rhizosphere Soil. Curr. Microbiol. 2021, 78, 218–228. [Google Scholar] [CrossRef]
- Hu, D.; Zang, Y.; Mao, Y.; Gao, B. Identification of Molecular Markers That Are Specific to the Class Thermoleophilia. Front. Microbiol. 2019, 10, 1185. [Google Scholar] [CrossRef]
- Weiss, B.; Souza, A.C.O.; Constancio, M.T.L.; Alvarenga, D.O.; Pylro, V.S.; Alves, L.M.C.; Varani, A.M. Unraveling a Lignocellulose-Decomposing Bacterial Consortium from Soil Associated with Dry Sugarcane Straw by Genomic-Centered Metagenomics. Microorganisms 2021, 9, 995. [Google Scholar] [CrossRef] [PubMed]
- Goss-Souza, D.; Mendes, L.W.; Borges, C.D.; Baretta, D.; Tsai, S.M.; Rodrigues, J.L.M. Soil microbial community dynamics and assembly under long-term land use change. FEMS Microbiol. Ecol. 2017, 93, 10. [Google Scholar] [CrossRef]
- Schmidt, R.; Gravuer, K.; Bossange, A.V.; Mitchell, J.; Scow, K. Long-term use of cover crops and no-till shift soil microbial community life strategies in agricultural soil. PLoS ONE 2018, 13, e0192953. [Google Scholar] [CrossRef]
- Liu, C.; Li, L.; Xie, J.; Coulter, J.A.; Zhang, R.; Luo, Z.; Cai, L.; Wang, L.; Gopalakrishnan, S. Soil Bacterial Diversity and Potential Functions Are Regulated by Long-Term Conservation Tillage and Straw Mulching. Microorganisms 2020, 8, 836. [Google Scholar] [CrossRef] [PubMed]
- Behnke, G.D.; Kim, N.; Zabaloy, M.C.; Riggins, C.W.; Rodriguez-Zas, S.; Villamil, M.B. Soil Microbial Indicators within Rotations and Tillage Systems. Microorganisms 2021, 9, 1244. [Google Scholar] [CrossRef]
- Duan, N.L.; Li, L.; Liang, X.; Fine, A.; Zhuang, J.; Radosevich, M.; Schaeffer, S.M. Variation in Bacterial Community Structure under Long-Term Fertilization, Tillage, and Cover Cropping in Continuous Cotton Production. Front. Microbiol. 2022, 13, 847005. [Google Scholar] [CrossRef] [PubMed]
- Firth, A.G.; Brooks, J.P.; Locke, M.A.; Morin, D.J.; Brown, A.; Baker, B.H. Soil bacterial community dynamics in plots managed with cover crops and no-till farming in the Lower Mississippi Alluvial Valley, USA. J. Appl. Microbiol. 2023, 134, lxac051. [Google Scholar] [CrossRef]
- Sengupta, A.; Dick, W.A. Bacterial Community Diversity in Soil under two Tillage Practices as Determined by Pyrosequencing. Microb. Ecol. 2015, 70, 853–859. [Google Scholar] [CrossRef]
- Frøslev, T.G.; Nielsen, I.B.; Santos, S.S.; Barnes, C.J.; Bruun, H.H.; Ejrnæs, R. The biodiversity effect of reduced tillage on soil microbiota. Ambio 2022, 51, 1022–1033. [Google Scholar] [CrossRef]
- Kerdraon, L.; Balesdent, M.H.; Barret, M.; Laval, V.; Suffert, F. Crop Residues in Wheat-Oilseed Rape Rotation System: A Pivotal, Shifting Platform for Microbial Meetings. Microb. Ecol. 2019, 77, 931–945. [Google Scholar] [CrossRef]
- Naumova, N.B.; Belanov, I.P.; Alikina, T.Y.; Kabilov, M.R. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Syst. 2021, 5, 14. [Google Scholar] [CrossRef]
- Will, C.; Thürmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniel, R. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [PubMed]
- Del Montero-Calasanz, M.C.; Meier-Kolthoff, J.P.; Zhang, D.-F.; Yaramis, A.; Rohde, M.; Woyke, T.; Kyrpides, N.C.; Schumann, P.; Li, W.-J.; Göker, M. Genome-Scale Data Call for a Taxonomic Rearrangement of Geodermatophilaceae. Front. Microbiol. 2017, 8, 2501. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Zeng, Q.C.; Huang, Y.M.; Ni, Y.X. Effects of the Farmland-to-Forest/Grassland Conversion Program on the Soil Bacterial Community in the Loess Hilly. Region. Huan Jing Ke Xue 2018, 39, 1824–1832. (In Chinese) [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.; Jo, H.Y.; Finneran, K.T.; Kwon, M.J. Diversity and composition of soil Acidobacteria and Proteobacteria communities as a bacterial indicator of past land-use change from forest to farmland. Sci. Total Environ. 2021, 797, 148944. [Google Scholar] [CrossRef]
- Yan, Y.; Sun, X.; Sun, F.; Zhao, Y.; Sun, W.; Guo, J.; Zhang, T. Sensitivity of soil fungal and bacterial community compositions to nitrogen and phosphorus additions in a temperate meadow. Plant Soil 2022, 471, 477–490. [Google Scholar] [CrossRef]
- Liu, J.J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Cai, L.; Guo, Z.; Zhang, J.; Gai, Z.; Liu, J.; Meng, Q.; Liu, X. No tillage and residue mulching method on bacterial community diversity regulation in a black soil region of Northeastern China. PLoS ONE 2021, 16, e0256970. [Google Scholar] [CrossRef]
- Gabbarini, L.A.; Figuerola, E.; Frene, J.P.; Robledo, N.B.; Ibarbalz, F.M.; Babin, D.; Smalla, K.; Erijman, L.; Wall, L.G. Impacts of switching tillage to no-tillage and vice versa on soil structure, enzyme activities and prokaryotic community profiles in Argentinean semi-arid soils. FEMS Microbiol. Ecol. 2021, 97, fiab025. [Google Scholar] [CrossRef]
- Figuerola, E.L.; Guerrero, L.D.; Rosa, S.M.; Simonetti, L.; Duval, M.E.; Galantini, J.A.; Bedano, J.C.; Wall, L.G.; Erijman, L. Bacterial indicator of agricultural management for soil under no-till crop production. PLoS ONE 2012, 7, e51075. [Google Scholar] [CrossRef]
- Brunbjerg, A.K.; Bruun, H.H.; Dalby, L.; Classen, A.T.; Fløjgaard, C.; Frøslev, T.G.; Pryds Hansen, O.L.; Høye, T.T.; Moeslund, J.E.; Svenning, J.-C.; et al. Multi-taxon inventory reveals highly consistent biodiversity responses to ecospace variation. Oikos 2020, 129, 1381–1392. [Google Scholar] [CrossRef]
- Martínez Arbas, S.; Busi, S.B.; Queirós, P.; de Nies, L.; Herold, M.; May, P.; Wilmes, P.; Muller, E.E.L.; Narayanasamy, S. Challenges, Strategies, and Perspectives for Reference-Independent Longitudinal Multi-Omic Microbiome Studies. Front. Genet. 2021, 12, 666244. [Google Scholar] [CrossRef]
- Brown, S.P.; Veach, A.M.; Rigdon-Huss, A.R.; Grond, K.; Lickteig, S.K.; Lothamer, K.; Oliver, A.K.; Jumpponen, A. Scraping the bottom of the barrel: Are rare high throughput sequences artifacts? Fungal Ecol. 2015, 13, 221–225. [Google Scholar] [CrossRef]
- Brunbjerg, A.K.; Bruun, H.H.; Brøndum, L.; Classen, A.T.; Dalby, L.; Fog, K.; Frøslev, T.G.; Goldberg, I.; Hansen, A.J.; Hansen, M.D.D.; et al. A systematic survey of regional multi-taxon biodiversity: Evaluating strategies and coverage. BMC Ecol. 2019, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Dahllöf, I.; Baillie, H.; Kjelleberg, S. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 2000, 66, 3376–3380. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Cheng, J.; Mei, L.J.; Hu, J.; Piao, Z.; Yin, S.X. Multiple copies of 16s rRNA gene affect the restriction patterns and DGGE profile as revealed by analysis of genome database. Mikrobiologiia 2010, 79, 664–671. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Sun, D.-L.; Jiang, X.; Wu, Q.; Zhou, N.-Y. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 2013, 79, 5962–5969. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef]
- Ibal, J.C.; Pham, H.Q.; Park, C.E.; Shin, J.H. Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PLoS ONE 2019, 14, e0212090. [Google Scholar] [CrossRef]
- Lin, J.N.; Lai, C.H.; Lin, S.Y.; Lee, C.C.; Lee, N.Y.; Liu, P.Y.; Yang, C.H.; Huang, Y.H. Effect of Intragenomic Sequence Heterogeneity among Multiple 16S rRNA Genes on Species Identification of Elizabethkingia. Microbiol. Spectr. 2022, 10, e0133822. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 2003, 228, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Heisenberg, W. The Representation of Nature in Contemporary Physics. Daedalus 1958, 87, 95–108. Available online: http://www.jstor.org/stable/20026454 (accessed on 13 September 2023).
- Starke, R.; Pylro, V.S.; Morais, D.K. 16S rRNA Gene Copy Number Normalization does not Provide More Reliable Conclusions in Metataxonomic Surveys. Microb. Ecol. 2021, 81, 535–539. [Google Scholar] [CrossRef] [PubMed]

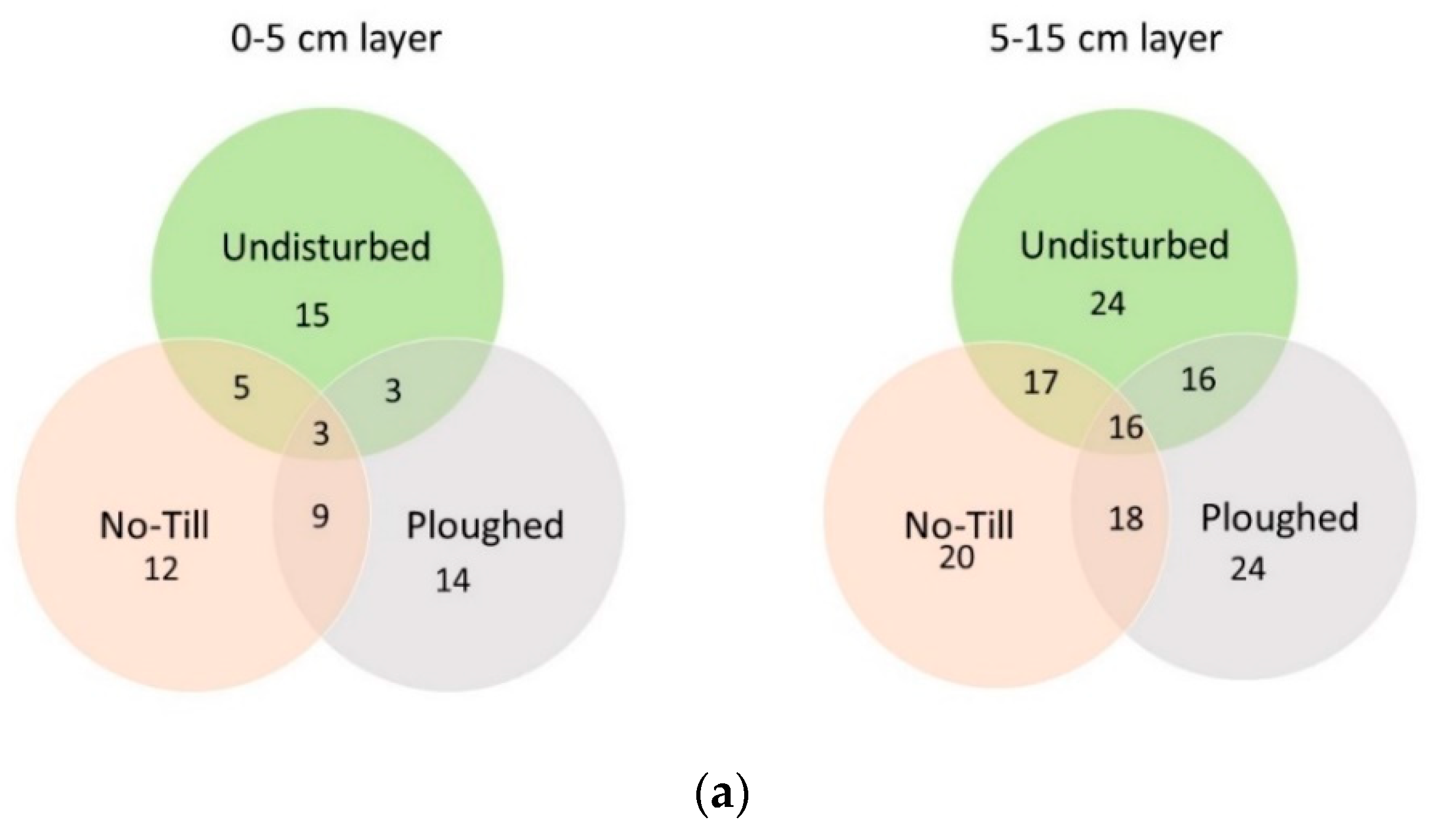
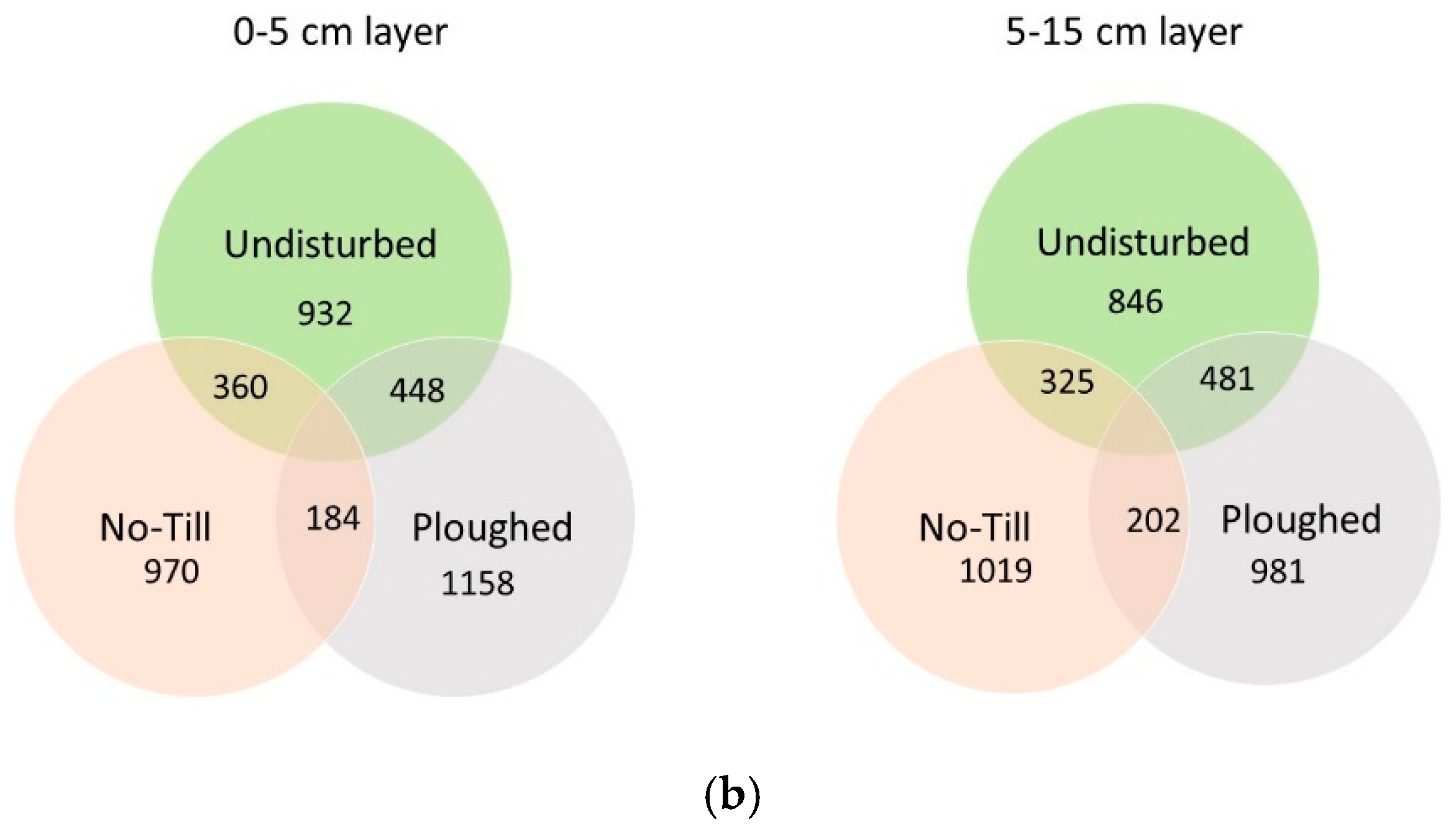
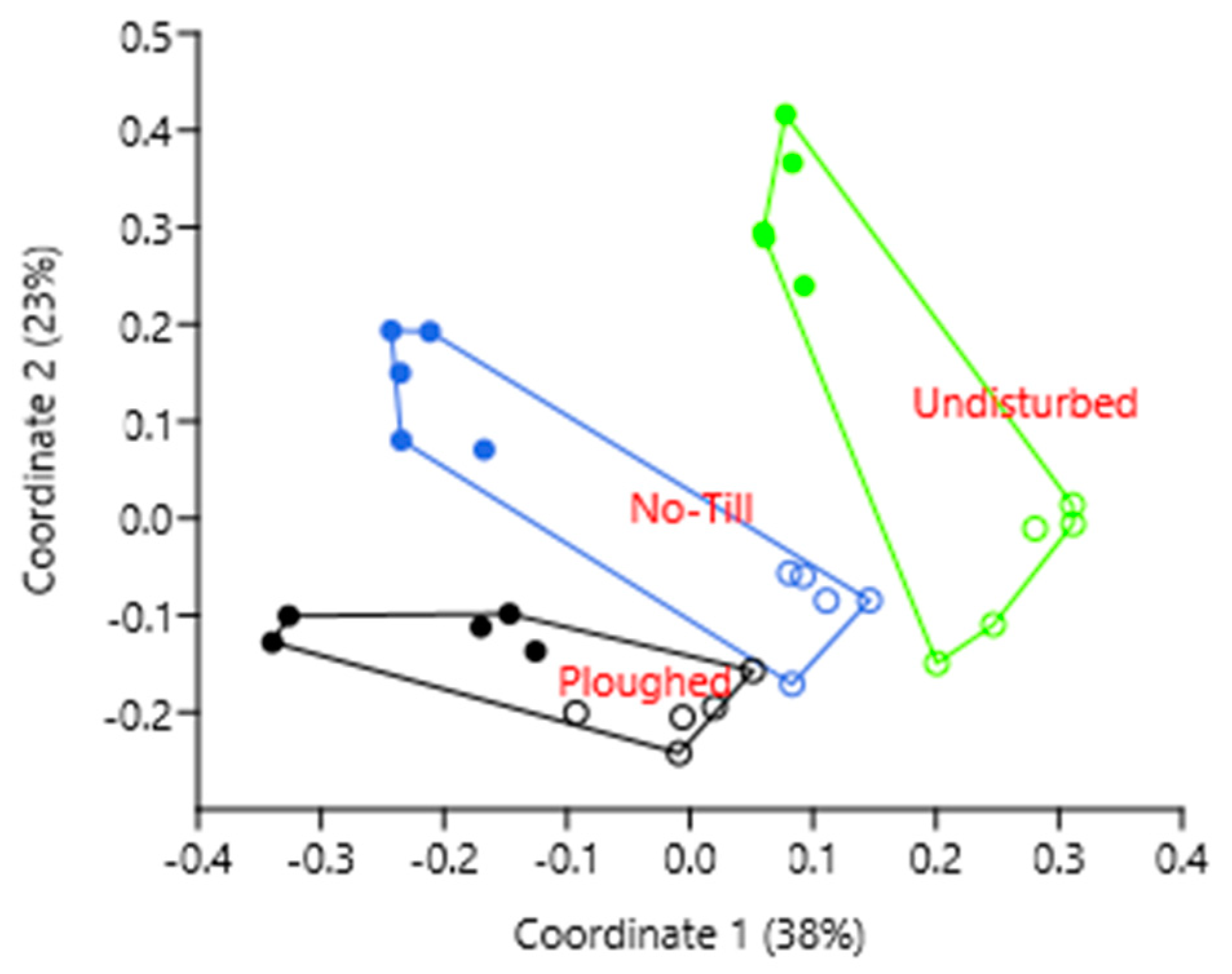
| Taxon | Undisturbed | Ploughed | No Till | |||
|---|---|---|---|---|---|---|
| 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | |
| Phylum level | ||||||
| Actinobacteria | 48.2 b 1 | 58.1 d | 38.0 a | 43.5 ab | 47.5 bc | 50.5 c |
| Acidobacteria | 19.6 a | 18.6 a | 32.9 b | 32.4 b | 18.7 a | 24.6 a |
| Proteobacteria | 12.7 b | 6.0 a | 9.3 ab | 5.9 a | 12.2 b | 6.3 a |
| Gemmatimonadetes | 1.9 a | 2.3 ab | 4.4 c | 4.3 c | 6.6 d | 3.5 bc |
| Chloroflexi | 1.3 a | 1.1 a | 3.2 d | 2.5 bc | 2.8 cd | 2.1 b |
| Verrucomicrobia | 4.9 c | 3.7 bc | 1.3 a | 1.3 a | 1.0 a | 1.8 ab |
| Bacteroidetes | 1.6 c | 0.3 a | 0.8 b | 0.4 a | 1.6 c | 0.5 ab |
| Class level | ||||||
| Thermoleophilia | 14.2 a | 29.1 c | 13.6 a | 20.4 b | 11.5 a | 20.6 b |
| Acidobacteria Gp6 | 10.9 ab | 7.1 a | 12.4 b | 14.8 b | 7.3 a | 13.6 b |
| Acidobacteria Gp16 | 2.1 a | 5.5 b | 12.4 c | 10.4 c | 4.6 ab | 4.3 ab |
| Actinobacteria | 22.0 c | 12.5 a | 14.0 b | 10.1 a | 24.5 c | 13.9 b |
| Alphaproteobacteria | 8.5 c | 5.0 ab | 5.4 ab | 4.0 a | 7.2 b | 4.6 ab |
| Spartobacteria | 4.6 b | 3.5 b | 1.0 a | 1.1 a | 0.7 a | 1.6 b |
| Rubrobacteria | 3.7 abc | 2.2 ab | 6.8 c | 4.4 b | 5.7 c | 3.1 ab |
| Acidobacteria | 0.4 a | 0.3 a | 3.1 cd | 2.8 c | 2.1 bc | 1.6 b |
| Gemmatimonadetes | 1.8 b | 0.6 a | 3.0 c | 1.8 b | 4.4 d | 1.6 b |
| Acidimicrobiia | 2.6 ab | 2.4 ab | 2.0 a | 2.4 ab | 2.0 a | 3.0 b |
| Acidobacteria Gp4 | 2.7 c | 2.9 c | 0.2 a | 0.4 a | 0.4 a | 1.5 b |
| Bacilli | 1.3 ab | 1.6 b | 0.2 a | 0.3 a | 0.5 a | 0.7 ab |
| Betaproteobacteria | 1.3 b | 0.3 a | 1.1 b | 0.5 a | 1.3 b | 0.4 a |
| Deltaproteobacteria | 2.2 d | 0.4 a | 1.7 c | 1.0 b | 2.4 d | 0.8 ab |
| Gammaproteobacteria | 0.7 b | 0.1 a | 0.9 c | 0.2 a | 1.2 c | 0.4 ab |
| Chitinophagia | 1.0 b | 0.2 a | 0.5 a | 0.3 a | 1.1 b | 0.4 a |
| Source | Sum of Squares | % In Variance | D.f. | Mean Square | F | p |
|---|---|---|---|---|---|---|
| Phylum level | ||||||
| Field | 1979.2 | 47 | 2 | 989.62 | 15.839 | 0.0001 |
| Layer | 543.91 | 13 | 1 | 543.91 | 8.7055 | 0.0018 |
| Interaction | 180.1 | 4 | 2 | 90.051 | 1.4413 | 0.2215 |
| Residual | 1499.5 | 36 | 24 | 62.479 | ||
| Total | 4202.7 | 100 | 29 | |||
| Class level | ||||||
| Field | 1271.2 | 27 | 2 | 635.61 | 10.515 | 0.0001 |
| Layer | 1689.8 | 35 | 1 | 1689.8 | 27.956 | 0.0001 |
| Interaction | 357.29 | 7 | 2 | 178.64 | 2.9554 | 0.0091 |
| Residual | 1450.7 | 30 | 24 | 60.445 | ||
| Total | 4769 | 100 | 29 | |||
| Order level | ||||||
| Field | 1295.3 | 31 | 2 | 647.67 | 11.778 | 0.0001 |
| Layer | 1185.7 | 28 | 1 | 1185.7 | 21.562 | 0.0001 |
| Interaction | 360.25 | 9 | 2 | 180.12 | 3.2755 | 0.002 |
| Residual | 1319.8 | 32 | 24 | 54.992 | ||
| Total | 4161.1 | 100 | 29 | |||
| Family level | ||||||
| Field | 1285.6 | 31 | 2 | 642.82 | 12.224 | 0.0001 |
| Layer | 1190 | 29 | 1 | 1190 | 22.631 | 0.0001 |
| Interaction | 362.79 | 9 | 2 | 181.4 | 3.4496 | 0.0012 |
| Residual | 1262.1 | 31 | 24 | 52.586 | ||
| Total | 4100.5 | 100 | 29 | |||
| Genus level | ||||||
| Field | 1270.4 | 32 | 2 | 635.2 | 12.198 | 0.0001 |
| Layer | 1112.6 | 28 | 1 | 1112.6 | 21.365 | 0.0001 |
| Interaction | 331.84 | 8 | 2 | 165.92 | 3.1862 | 0.0028 |
| Residual | 1249.8 | 32 | 24 | 52.073 | ||
| Total | 3964.6 | 100 | 29 | |||
| OTUs level | ||||||
| Field | 433.33 | 25 | 2 | 216.66 | 6.3671 | 0.0001 |
| Layer | 385.68 | 22 | 1 | 385.68 | 11.334 | 0.0001 |
| Interaction | 123.89 | 7 | 2 | 61.944 | 1.8203 | 0.0386 |
| Residual | 816.69 | 46 | 24 | 34.029 | ||
| Total | 1759.6 | 100 | 29 | |||
| Genus | Undisturbed | Ploughed | No Till | |||
|---|---|---|---|---|---|---|
| 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | |
| Actinoplanes | 2.6 d, 1 | 0.0 a | 0.4 b | 0.0 a | 1.5 c | 0.1 ab |
| Bacillus | 1.0 b | 1.1 b | 0.1 a | 0.0 a | 0.2 ab | 0.3 ab |
| Blastococcus | 0.0 a | 0.0 a | 3.3 c | 1.4 b | 4.7 d | 1.5 b |
| Gaiella | 1.5 a | 13.9 d | 6.2 b | 11.7 c | 2.9 a | 10.2 c |
| Acidobacteria_Gp4 | 2.9 c | 2.9 c | 0.4 a | 0.5 a | 0.5 a | 1.6 b |
| Acidobacteria_Gp6 | 11.0 ab | 7.1 a | 12.4 b | 14.9 b | 7.3 a | 13.6 b |
| Acidobacteria_Gp16 | 2.1 a | 5.6 b | 13.0 c | 10.9 c | 4.7 ab | 4.4 ab |
| Kribbella | 1.0 b | 1.0 b | 0.4 a | 0.5 ab | 0.9 b | 0.8 ab |
| Microlunatus | 2.5 a | 4.1 ab | 1.9 a | 2.4 a | 5.1 b | 4.1 ab |
| Pseudonocardia | 3.2 c | 0.3 a | 0.7 a | 0.6 a | 2.0 b | 0.8 a |
| Rubrobacter | 3.7 ab | 2.2 a | 6.8 b | 4.4 ab | 5.7 b | 3.1 a |
| Solirubrobacter | 0.8 a | 1.5 a | 1.4 a | 1.3 a | 1.8 a | 1.0 a |
| Spartobacteria_gis | 4.6 b | 3.5 b | 0.9 a | 1.1 a | 0.7 a | 1.5 a |
| un. 2 Acidimicrobiales | 0.9 b | 1.4 c | 0.4 a | 1.0 b | 0.4 a | 1.4 c |
| un. Acidobacteria | 0.8 a | 0.7 a | 3.7 c | 3.4 bc | 3.1 b | 2.3 b |
| un. Actinobacteria | 9.2 c | 16.7 e | 3.0 a | 9.0 c | 5.5 b | 13.1 d |
| un. Chloroflexi | 0.4 a | 0.5 ab | 1.5 d | 1.0 c | 1.3 d | 0.7 b |
| un. Gemmatimonadaceae | 1.4 b | 0.5 a | 2.7 c | 1.7 b | 3.7 d | 1.5 b |
| un. Gemmatimonadetes | 0.1 a | 1.7 b | 1.3 ab | 2.5 b | 2.1 b | 1.9 b |
| un. Micromonosporaceae | 2.3 c | 0.9 b | 0.4 a | 0.5 ab | 0.6 ab | 0.8 b |
| un. Rhizobiales | 4.3 b | 4.2 b | 1.5 a | 1.8 a | 0.9 a | 2.3 a |
| un. Solirubrobacterales | 9.1 c | 8.9 c | 4.8 a | 5.5 ab | 5.8 ab | 7.0 b |
| un. Thermoleophilia | 2.6 c | 4.6 d | 1.0 a | 1.8 b | 0.8 a | 2.3 bc |
| Index | Undisturbed | Ploughed | No Till | |||
|---|---|---|---|---|---|---|
| 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | 0–5 cm | 5–15 cm | |
| OTU richness | 932 ab, 1 | 846 a | 1158 b | 981 ab | 970 ab | 1019 ab |
| Chao-1 | 1349 ab | 1160 a | 1562 b | 1415 ab | 1402 ab | 1399 ab |
| Simpson (S) | 0.989 b | 0.985 a | 0.986 ab | 0.988 ab | 0.987 ab | 0.988 ab |
| Shannon’s | 5.55 b | 5.11 a | 5.51 b | 5.32 ab | 5.47 b | 5.40 b |
| Evenness | 0.28 c | 0.20 a | 0.22 ab | 0.21 a | 0.25 b | 0.22 ab |
| Equitability | 0.81 d | 0.76 a | 0.79 bc | 0.77 ab | 0.80 c | 0.78 b |
| Berger-Parker | 0.05 a | 0.05 a | 0.07 b | 0.05 ab | 0.06 ab | 0.05 a |
| Dominance (1-S) | 0.011 a | 0.015 b | 0.014 ab | 0.012 ab | 0.013 ab | 0.012 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumova, N.; Barsukov, P.; Baturina, O.; Rusalimova, O.; Kabilov, M. West-Siberian Chernozem: How Vegetation and Tillage Shape Its Bacteriobiome. Microorganisms 2023, 11, 2431. https://doi.org/10.3390/microorganisms11102431
Naumova N, Barsukov P, Baturina O, Rusalimova O, Kabilov M. West-Siberian Chernozem: How Vegetation and Tillage Shape Its Bacteriobiome. Microorganisms. 2023; 11(10):2431. https://doi.org/10.3390/microorganisms11102431
Chicago/Turabian StyleNaumova, Natalia, Pavel Barsukov, Olga Baturina, Olga Rusalimova, and Marsel Kabilov. 2023. "West-Siberian Chernozem: How Vegetation and Tillage Shape Its Bacteriobiome" Microorganisms 11, no. 10: 2431. https://doi.org/10.3390/microorganisms11102431





