Dysbiosis by Eradication of Helicobacter pylori Infection Associated with Follicular Gastropathy and Pangastropathy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characterization of the Gastric Microenvironment
2.1.1. Study Subjects and Sample Collection
2.1.2. Histopathological Evaluation
2.1.3. Isolation and Characterization of H. pylori Cultures
Antibiotic Susceptibility Profiling and Molecular Genotyping
2.1.4. NGS Sequencing
Metagenomic Analysis: Sequencing Data Quality Control and Processing
2.2. Correlation of the Presence of C. acnes and H. pylori with Gastric Diseases
2.2.1. Study Subjects and Sample Collection
2.2.2. FFPE Deparaffinization for RNA Isolation
2.2.3. Quantitative Real-Time PCR (qRT-PCR)
2.2.4. Validation of qRT-PCR, Data Treatment and Statistical Analysis
3. Results
3.1. Characterization of the Gastric Microenvironment
3.1.1. Histopathological Evaluation
3.1.2. Molecular Identification of H. pylori and Urease Detection
3.1.3. Antibiotic Susceptibility Profiling and Molecular Genotyping
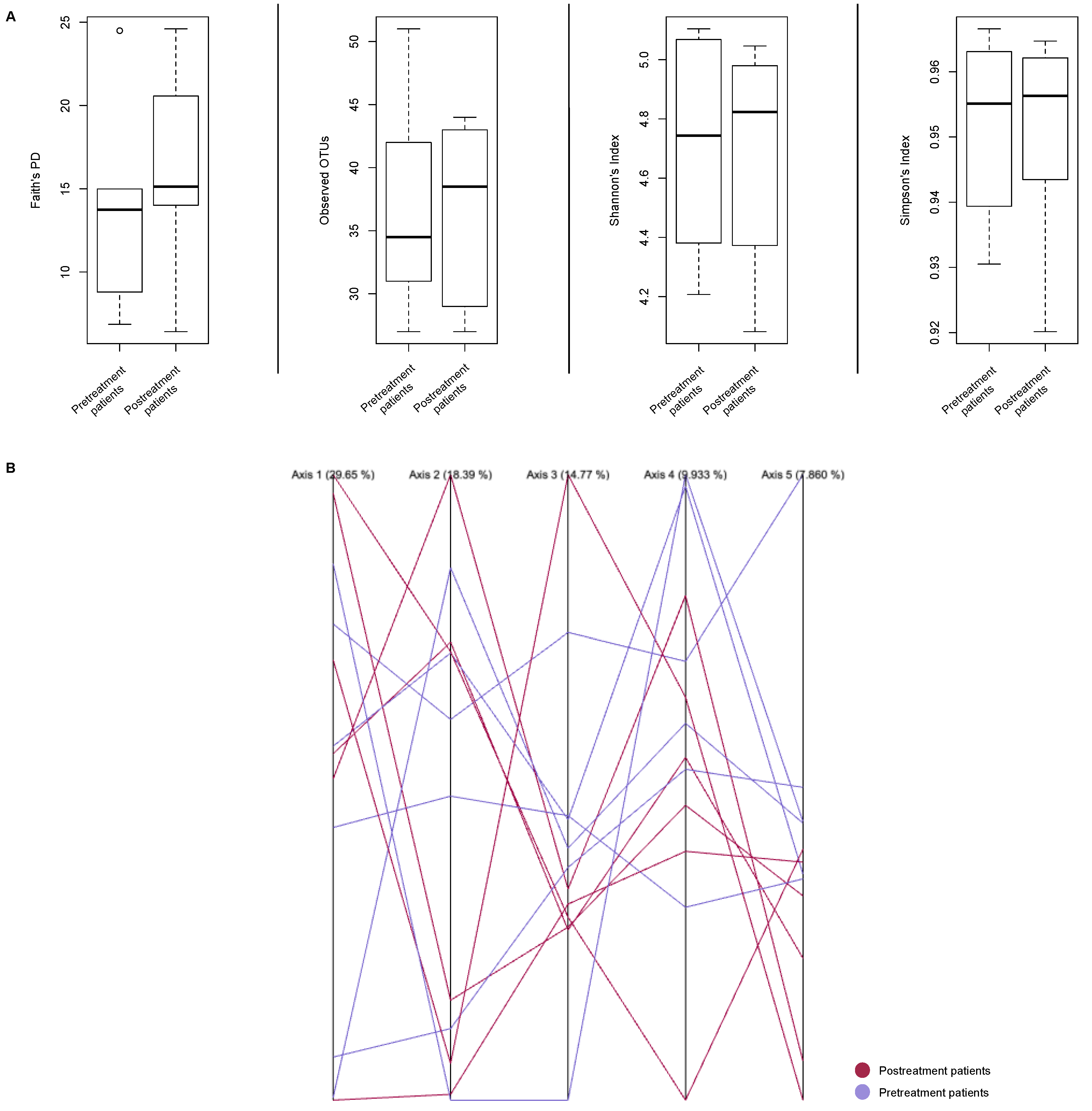
3.1.4. Metagenomic Analysis
3.2. Correlation of C. acnes in Gastric Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.L.; Lollie, T.K.; Chen, Z.; Narasimhalu, T.; Wang, H.L. Histopathologic diagnosis of gastritis and gastropathy: A narrative review. Dig. Med. Res. 2022, 6, 3. [Google Scholar] [CrossRef]
- Carlosama-Rosero, Y.; Bolaños-Bravo, H.; Sierra-Tórres, C.; Rosero, E. Asociación de los genotipos cagA, vacA e IceA de H. pylori con la gastritis crónica y folicular en una población colombiana con alto riesgo de cáncer gástrico. Rev. Gastroenterol. Mex. 2019, 84, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Kayacetin, S.; Guresci, S. What is gastritis? What is gastropathy? How is it classified? Turk. J. Gastroenterol. Off. J. Turk. Soc. Gastroenterol. 2014, 25, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Atherton, J.C.; Blaser, M.J. Coadaptation of Helicobacter pylori and humans: Ancient history, modern implications. J. Clin. Investig. 2009, 119, 2475–2487. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-C. Treatment of Helicobacter pylori infection: Current status and future concepts. World J. Gastroenterol. 2014, 20, 5283–5293. [Google Scholar] [CrossRef]
- Bielsa-Fernández, M.; la Cuesta, J.T.-D.; Lizárraga-López, J.; Remes-Troche, J.; Carmona-Sánchez, R.; Aldana-Ledesma, J.; Avendaño-Reyes, J.; Ballesteros-Amozorrutia, M.; De Ariño, M.; de Giau-Triulzi, L.; et al. Consenso mexicano sobre diagnóstico, prevención y tratamiento de la gastropatía y enteropatía por antiinflamatorios no esteroideos. Rev. Gastroenterol. Mex. 2020, 85, 190–206. [Google Scholar] [CrossRef]
- Lanza, F.L.; Chan, F.K.; Quigley, E.M.; Practice Parameters Committee of the American College of Gastroenterology. Guidelines for Prevention of NSAID-Related Ulcer Complications. Am. J. Gastroenterol. 2009, 104, 728–738. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- National Institute of Diabetes and Digestive and Kidney Diseases. Gastritis & Gastropathy. Available online: https://www.niddk.nih.gov/health-information/digestive-diseases/gastritis-gastropathy (accessed on 12 October 2023).
- Kato, T.; Yagi, N.; Kamada, T.; Shimbo, T.; Watanabe, H.; Ida, K.; Study Group for Establishing Endoscopic Diagnosis of Chronic Gastritis. Diagnosis of Helicobacter Pylori Infection in Gastric Mucosa by Endoscopic Features: A Multicenter Prospective Study. Dig. Endosc. Off. J. Jpn. Gastroenterol. Endosc. Soc. 2013, 25, 508–518. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.-M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef]
- Ladrón-De-Guevara, L.; Bornstein-Quevedo, L.; González-Huezo, S.; Castañeda-Romero, B.; Costa, F.; di Silvio-López, M. Erradicación de Helicobacter pylori en México con un esquema basado en levofloxacina versus la triple terapia estándar: Resultados de un estudio clínico de fase iiib, abierto, aleatorizado, de no inferioridad. Rev. Gastroenterol. Mex. 2018, 84, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Guía de Práctica Clínica Diagnóstico y Tratamiento de la Dispepsia Funcional. 2009. México. Instituto Mexicano del Seguro Social. Available online: http://www.imss.gob.mx/sites/all/statics/guiasclinicas/071GER.pdf (accessed on 15 October 2022).
- Wasielica-Berger, J.; Gugnacki, P.; Mlynarczyk, M.; Rogalski, P.; Swidnicka-Siergiejko, A.; Antonowicz, S.; Krzyzak, M.; Maslach, D.; Dabrowski, A.; Daniluk, J. Comparative Effectiveness of Various Eradication Regimens for Helicobacter Pylori Infection in the Northeastern Region of Poland. Int. J. Environ. Res. Public Health 2022, 19, 6921. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ramirez, U.; Valencia-Mayoral, P.; Mendoza-Elizalde, S.; Murillo-Eliosa, J.R.; Santos, F.S.; Contreras-Rodríguez, A.; Zúñiga, G.; Aguilar-Rodea, P.; Jiménez-Rojas, V.L.; Galindo, J.C.V.; et al. Role of Helicobacter pylori and Other Environmental Factors in the Development of Gastric Dysbiosis. Pathogens 2021, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Honggang, Y. The Role of Non-H. Pylori Bacteria in the Development of Gastric Cancer. Am. J. Cancer Res. 2020, 10, 2271–2281. [Google Scholar] [PubMed]
- Petra, C.V.; Rus, A.; Dumitrașcu, D.L. Gastric Microbiota: Tracing the Culprit. Med. Pharm. Rep. 2017, 90, 369–376. [Google Scholar] [CrossRef]
- Boisrenoult, P. Cutibacterium acnes prosthetic joint infection: Diagnosis and treatment. Orthop. Traumatol. Surg. Res. 2018, 104, S19–S24. [Google Scholar] [CrossRef]
- Claesen, J.; Spagnolo, J.B.; Ramos, S.F.; Kurita, K.L.; Byrd, A.L.; Aksenov, A.A.; Melnik, A.V.; Wong, W.R.; Wang, S.; Hernandez, R.D.; et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 2020, 12, eaay5445. [Google Scholar] [CrossRef]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.J.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two Major Sentinels of Skin Microbiota and the Influence of Cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef]
- Delgado, S.; Suárez, A.; Mayo, B. Identification, typing and characterisation of Propionibacterium strains from healthy mucosa of the human stomach. Int. J. Food Microbiol. 2011, 149, 65–72. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, H.; Salar-Vidal, L.; Gollnick, H.P.M.; Lood, R. A Janus-Faced Bacterium: Host-Beneficial and -Detrimental Roles of Cutibacterium acnes. Front. Microbiol. 2021, 12, 673845. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Arques, A.; Wurm, P.; Trajanoski, S.; Schauer, S.; Kienesberger, S.; Halwachs, B.; Högenauer, C.; Langner, C.; Gorkiewicz, G. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 2016, 240, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2018, 40, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and Grading of Gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Wayne, P.A., Ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2015. [Google Scholar]
- Mendoza-Elizalde, S.; Arteaga-Resendiz, N.K.; Valencia-Mayoral, P.; Luna, R.C.; Moreno-Espinosa, S.; Arenas-Huertero, F.; Zúñiga, G.; Velázquez-Guadarrama, N. Diversification of the vacAs1m1 and vacAs2m2 Strains of Helicobacter pylori in Meriones unguiculatus. Front. Microbiol. 2016, 7, 1758. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.r-project.org/ (accessed on 15 October 2022).
- Miura, Y.; Ishige, I.; Soejima, N.; Suzuki, Y.; Uchida, K.; Kawana, S.; Eishi, Y. Quantitative PCR of Propionibacterium acnes DNA in samples aspirated from sebaceous follicles on the normal skin of subjects with or without acne. J. Med. Dent. Sci. 2010, 57, 65–74. [Google Scholar]
- Roshdy, T.M.; Saleh, S.A.; Abass, N.H.; Bassiouny, K.; Khalil, H. Disturbing intracellular replication of Helicobacter pylori by sorafenib treatment in-vitro. Afr. J. Biol. Sci. 2021, 17, 285–295. [Google Scholar] [CrossRef]
- Abadi, A.T.B. Diagnosis of Helicobacter pylori Using Invasive and Noninvasive Approaches. J. Pathog. 2018, 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ailloud, F.; Didelot, X.; Woltemate, S.; Pfaffinger, G.; Overmann, J.; Bader, R.C.; Schulz, C.; Malfertheiner, P.; Suerbaum, S. Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps. Nat. Commun. 2019, 10, 2273. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.G.; Choi, I.J.; Lee, J.Y.; Cho, S.-J.; Nam, B.-H.; Kook, M.-C.; Hong, E.K.; Kim, Y.-W. Biopsy site for detecting Helicobacter pylori infection in patients with gastric cancer. J. Gastroenterol. Hepatol. 2009, 24, 469–474. [Google Scholar] [CrossRef]
- Noh, C.-K.; Lee, G.H.; Park, J.W.; Roh, J.; Han, J.H.; Lee, E.; Park, B.; Lim, S.G.; Shin, S.J.; Cheong, J.Y.; et al. Diagnostic accuracy of “sweeping” method compared to conventional sampling in rapid urease test for Helicobacter pylori detection in atrophic mucosa. Sci. Rep. 2020, 10, 18483. [Google Scholar] [CrossRef]
- Ierardi, E.; Losurdo, G.; Mileti, A.; Paolillo, R.; Giorgio, F.; Principi, M.; Di Leo, A. The Puzzle of Coccoid Forms of Helicobacter pylori: Beyond Basic Science. Antibiotics 2020, 9, 293. [Google Scholar] [CrossRef]
- Pennelli, G.; Grillo, F.; Galuppini, F.; Ingravallo, G.; Pilozzi, E.; Rugge, M.; Fiocca, R.; Fassan, M.; Mastracci, L. Gastritis: Update on etiological features and histological practical approach. Pathologica 2020, 112, 153–165. [Google Scholar] [CrossRef]
- Fucarino, A.; Burgio, S.; Paladino, L.; Bavisotto, C.C.; Pitruzzella, A.; Bucchieri, F.; Cappello, F. The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure. Anatomia 2022, 1, 186–203. [Google Scholar] [CrossRef]
- Nakamura, H.; Yoshiyama, H.; Takeuchi, H.; Mizote, T.; Okita, K.; Nakazawa, T. Urease Plays an Important Role in the Chemotactic Motility of Helicobacter pylori in a Viscous Environment. Infect. Immun. 1998, 66, 4832–4837. [Google Scholar] [CrossRef]
- Brandi, G.; Biavati, B.; Calabrese, C.; Granata, M.; Nannetti, A.; Mattarelli, P.; Di Febo, G.; Saccoccio, G.; Biasco, G. Urease-Positive Bacteria Other than Helicobacter pylori in Human Gastric Juice and Mucosa. Am. J. Gastroenterol. 2006, 101, 1756–1761. [Google Scholar] [CrossRef]
- Ramírez-Lázaro, M.J.; Lario, S.; Calvet, X.; Sánchez-Delgado, J.; Montserrat, A.; Quílez, E.M.; Casalots, A.; Suarez, D.; Campo, R.; Brullet, E.; et al. Occult H. pylori infection partially explains ‘false-positive’ results of 13 C-urea breath test. United Eur. Gastroenterol. J. 2015, 3, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Non, H. Pylori Bacteria with Urease Activity Identified in the Human Stomach. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 605. [Google Scholar] [CrossRef]
- Arama, S.S.; Tiliscan, C.; Negoita, C.; Croitoru, A.; Arama, V.; Mihai, C.M.; Pop, F.; Garg, A. Efficacy of 7-Day and 14-Day Triple Therapy Regimens for the Eradication of Helicobacter pylori: A Comparative Study in a Cohort of Romanian Patients. Gastroenterol. Res. Pract. 2016, 2016, 5061640. [Google Scholar] [CrossRef] [PubMed]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Dingsdag, S.A.; Hunter, N. Metronidazole: An update on metabolism, structure–cytotoxicity and resistance mechanisms. J. Antimicrob. Chemother. 2017, 73, 265–279. [Google Scholar] [CrossRef]
- Rizvanov, A.A.; Haertlé, T.; Bogomolnaya, L.; Abadi, A.T.B. Helicobacter pylori and Its Antibiotic Heteroresistance: A Neglected Issue in Published Guidelines. Front. Microbiol. 2019, 10, 1796. [Google Scholar] [CrossRef]
- Boyanova, L.; Hadzhiyski, P.; Gergova, R.; Markovska, R. Evolution of Helicobacter pylori Resistance to Antibiotics: A Topic of Increasing Concern. Antibiotics 2023, 12, 332. [Google Scholar] [CrossRef]
- Kaklikkaya, N.; Cubukcu, K.; Aydin, F.; Bakir, T.; Erkul, S.; Tosun, I.; Topbas, M.; Yazici, Y.; Buruk, C.K.; Erturk, M. Significance of cagA status and vacA subtypes of Helicobacter pylori in determining gastric histopathology: Virulence markers of H. pylori and histopathology. J. Gastroenterol. Hepatol. 2006, 21, 1042–1047. [Google Scholar] [CrossRef]
- Liu, X.; Nie, W.; Liang, J.; Li, Y. Interaction of Helicobacter Pylori with Other Microbiota Species in the Development of Gastric Cancer. Arch. Clin. Microbiol. 2017, 8, 67. [Google Scholar] [CrossRef]
- Vital, J.S.; Tanoeiro, L.; Lopes-Oliveira, R.; Vale, F.F. Biomarker Characterization and Prediction of Virulence and Antibiotic Resistance from Helicobacter pylori Next Generation Sequencing Data. Biomolecules 2022, 12, 691. [Google Scholar] [CrossRef]
- Nogueira, C.; Figueiredo, C.; Carneiro, F.; Gomes, A.T.; Barreira, R.; Figueira, P.; Salgado, C.; Belo, L.; Peixoto, A.; Bravo, J.C.; et al. Helicobacter pylori Genotypes May Determine Gastric Histopathology. Am. J. Pathol. 2001, 158, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Roesler, B.M.; Rabelo-Gonçalves, E.M.; Zeitune, J.M. Virulence Factors of Helicobacter pylori: A Review. Clin. Med. Insights Gastroenterol. 2014, 7, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Demırtürk, L.; Özel, A.M.; Yazgan, Y.; Solmazgül, E.; Yildirim; Gültepe, M.; Gürbüz, A.K. CagA Status in Dyspeptic Patients with and without Peptic Ulcer Disease in Turkey: Association with Histopathologic Findings. Helicobacter 2001, 6, 163–168. [Google Scholar] [CrossRef]
- Norma, U.E.; Angelina, C.J.N.; Lilia, M.V.L.V.L.; Mayra, C.L.C.; Aurora, M.N.R.; Joseacute, A.H.H.N.; Juan, A.R.C.; Estela, R.B. Prevalence of Helicobacter pylori cagA and vacA genotypes in a population from Northeastern Mexico with chronic gastritis and intestinal metaplasia. Afr. J. Microbiol. Res. 2013, 7, 1409–1414. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound Alterations of Intestinal Microbiota following a Single Dose of Clindamycin Results in Sustained Susceptibility to Clostridium difficile-Induced Colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Duynhoven, Y.T.V.; Jonge, R.D. Transmission of Helicobacter Pylori: A Role for Food? Bull. World Health Organ. 2001, 79, 455–460. [Google Scholar]
- Akamatsu, T.; Tabata, K.; Hironga, M.; Kawakami, H.; Yyeda, M. Transmission of Helicobacter pylori infection via flexible fiberoptic endoscopy. Am. J. Infect. Control. 1996, 24, 396–401. [Google Scholar] [CrossRef]
- Kim, D.-H.; Son, B.K.; Min, K.-W.; Han, S.K.; Na, J.U.; Choi, P.C.; Kim, H.-L.; Kwon, M.J.; Oh, Y.H.; Jung, W.Y.; et al. Chronic Gastritis Is Associated with a Decreased High-Density Lipid Level: Histological Features of Gastritis Based on the Updated Sydney System. J. Clin. Med. 2020, 9, 1856. [Google Scholar] [CrossRef]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Birg, A.; Ritz, N.L.; Lin, H.C. The Unknown Effect of Antibiotic-Induced Dysbiosis on the Gut Microbiota. Microbiome Metabolome Diagn. Ther. Other Strateg. Appl. 2019, 195–200. [Google Scholar] [CrossRef]
- Reed, T.; Lushington, G.H.; Xia, Y.; Hirakawa, H.; Travis, D.M.; Mure, M.; Scott, E.E.; Limburg, J. Crystal Structure of Histamine Dehydrogenase from Nocardioides simplex. J. Biol. Chem. 2010, 285, 25782–25791. [Google Scholar] [CrossRef]
- Hagiya, H.; Sugiyama, J.; Kuroe, Y.; Nojima, H.; Naito, H.; Hagioka, S.; Morimoto, N.; Murase, T. Delftia acidovorans bacteremia caused by bacterial translocation after organophosphorus poisoning in an immunocompetent adult patient. J. Infect. Chemother. 2013, 19, 338–341. [Google Scholar] [CrossRef]
- Ghattargi, V.C.; Nimonkar, Y.S.; Sape, K.; Prakash, O.; Suryavanshi, M.V.; Shouche, Y.S.; Meti, B.S.; Pawar, S.P. Functional and Comparative Genomics of Niche-Specific Adapted Actinomycetes Kocuria rhizophila Strain D2 Isolated from Healthy Human Gut. bioRxiv 2018, 400242. [Google Scholar] [CrossRef]
- Kuda, T. Quality Improvement and Fermentation Control in Fish Products. In Advances in Fermented Foods and Beverages: Improving Quality, Technologies and Health Benefits; Woodhead Publishing: Cambridge, UK, 2015; pp. 377–390. [Google Scholar] [CrossRef]
- Gomi, A.; Harima-Mizusawa, N.; Shibahara-Sone, H.; Kano, M.; Miyazaki, K.; Ishikawa, F. Effect of Bifidobacterium bifidum BF-1 on gastric protection and mucin production in an acute gastric injury rat model. J. Dairy Sci. 2013, 96, 832–837. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef] [PubMed]
- Butta, H.; Sardana, R.; Vaishya, R.; Singh, K.N.; Mendiratta, L. Bifidobacterium: An Emerging Clinically Significant Metronidazole-resistant Anaerobe of Mixed Pyogenic Infections. Cureus 2017, 9, e1134. [Google Scholar] [CrossRef] [PubMed]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Simonsen, G.S.; Klingenberg, C. Bifidobacterium Bacteremia: Clinical Characteristics and a Genomic Approach To Assess Pathogenicity. J. Clin. Microbiol. 2017, 55, 2234–2248. [Google Scholar] [CrossRef]
- Segers, P.; Vancanneyt, M.; Pot, B.; Torck, U.; Hoste, B.; Dewettinck, D.; Falsen, E.; Kersters, K.; DE Vos, P. Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Busing, Doll, and Freytag 1953 in Brevundimonas gen. nov. as Brevundimonas diminuta comb. nov. and Brevundimonas vesicularis comb. nov., Respectively. Int. J. Syst. Evol. Microbiol. 1994, 44, 499–510. [Google Scholar] [CrossRef]
- Schwartz, M.A.; Tabet, S.R.; Collier, A.C.; Wallis, C.K.; Carlson, L.C.; Nguyen, T.T.; Kattar, M.M.; Coyle, M.B. Central Venous Catheter–Related Bacteremia Due to Tsukamurella Species in the Immunocompromised Host: A Case Series and Review of the Literature. Clin. Infect. Dis. 2002, 35, e72–e77. [Google Scholar] [CrossRef]
- Lee, M.; Srinivasan, S.; Kim, M.K. New taxa in Alphaproteobacteria: Brevundimonas olei sp. nov., an esterase-producing bacterium. J. Microbiol. 2010, 48, 616–622. [Google Scholar] [CrossRef]
- Kandi, V.; Palange, P.; Vaish, R.; Bhatti, A.B.; Kale, V.; Kandi, M.R.; Bhoomagiri, M.R. Emerging Bacterial Infection: Identification and Clinical Significance of Kocuria Species. Cureus 2016, 8, e731. [Google Scholar] [CrossRef] [PubMed]
- Alkanany, F.N.; Gmais, S.A.; Maki, A.A.; Altaee, A.M. Estimation of Bacterial Biodegradability of PAH in Khor Al-Zubair Channel, Southern Iraq. Int. J. Mar. Sci. 2017, 7, 42. [Google Scholar] [CrossRef]
- Wisplinghoff, H. Pseudomonas spp., Acinetobacter spp. and Miscellaneous Gram-Negative Bacilli. In Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1579–1599.e2. [Google Scholar] [CrossRef]
- Peduto, L.; Philippe, J.S.; Giulia, N. Chapter Three-Intestinal Stem Cells and Their Niche at Homeo-stasis and Under Stress. In Advances in Stem Cells and Their Niches; Bonnet, D., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 2, pp. 77–97. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp.: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, M.M.; Gent, J.F.; Kong, Y.; Halpin, A.L.; Pineles, L.; Harris, A.D.; Johnson, J.K. Gastrointestinal Microbiota Disruption and Risk of Colonization with Carbapenem-resistant Pseudomonas aeruginosa in Intensive Care Unit Patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018, 69, 604–613. [Google Scholar] [CrossRef]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Wagner, J.; Short, K.; Catto-Smith, A.G.; Cameron, D.J.S.; Bishop, R.F.; Kirkwood, C.D. Identification and Characterisation of Pseudomonas 16S Ribosomal DNA from Ileal Biopsies of Children with Crohn’s Disease. PLoS ONE 2008, 3, e3578. [Google Scholar] [CrossRef]
- Gao, Y.-D.; Zhao, Y.; Huang, J. Metabolic Modeling of Common Escherichia coli Strains in Human Gut Microbiome. BioMed Res. Int. 2014, 2014, 694967. [Google Scholar] [CrossRef]
- Conway, T.; Cohen, P.S. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiol. Spectr. 2015, 3, 343–362. [Google Scholar] [CrossRef]
- Sonnenborn, U. Escherichia coli strain Nissle 1917—From bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363, fnw212. [Google Scholar] [CrossRef]
- Fang, K.; Jin, X.; Hong, S.H. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci. Rep. 2018, 8, 4939. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Lemieszek, M.K.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. AAEM 2016, 23, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Büyükcam, A.; Tuncer, Ö.; Gür, D.; Sancak, B.; Ceyhan, M.; Cengiz, A.B.; Kara, A. Clinical and microbiological characteristics of Pantoea agglomerans infection in children. J. Infect. Public Health 2018, 11, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Delgado, S.; Cabrera-Rubio, R.; Mira, A.; Suárez, A.; Mayo, B. Microbiological Survey of the Human Gastric Ecosystem Using Culturing and Pyrosequencing Methods. Microb. Ecol. 2013, 65, 763–772. [Google Scholar] [CrossRef]
- Kovaleva, J.; Degener, J.E.; van der Mei, H.C. Methylobacterium and Its Role in Health Care-Associated Infection. J. Clin. Microbiol. 2014, 52, 1317–1321. [Google Scholar] [CrossRef]
- Mukherjee, S.; Ramesh, A. Bacteriocin-producing strains of Lactobacillus plantarum inhibit adhesion of Staphylococcus aureus to extracellular matrix: Quantitative insight and implications in antibacterial therapy. J. Med. Microbiol. 2015, 64, 1514–1526. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57 (Suppl. 1), 1–14. [Google Scholar] [CrossRef]
- Liu, J.; Xue, Y.; Zhou, L. Detection of gastritis-associated pathogens by culturing of gastric juice and mucosa. Int. J. Clin. Exp. Pathol. 2018, 11, 2214–2220. [Google Scholar]
- Barbosa, A.J.A.; Boldt, M.S.; Rodrigues, C.B.; Silva, C.S.C.; Ferreira, H.M.; Pereira, R.D. Histopathological features of mucosa atrophy in atrophic body gastritis. J. Bras. De Patol. E Med. Lab. 2016, 52, 50–54. [Google Scholar] [CrossRef]
- Jonaitis, P.; Kupcinskas, L.; Kupcinskas, J. Molecular Alterations in Gastric Intestinal Metaplasia. Int. J. Mol. Sci. 2021, 22, 5758. [Google Scholar] [CrossRef]
- Schubert, J.P.; Rayner, C.K.; Costello, S.P.; Roberts-Thomson, I.C.; Forster, S.C.; Bryant, R.V. Helicobacter pylori: Have potential benefits been overlooked? JGH Open 2022, 6, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Nordenstedt, H.; Graham, D.Y.; Kramer, J.R.; Rugge, M.; Verstovsek, G.; Fitzgerald, S.; Alsarraj, A.; Shaib, Y.; Velez, M.E.; Abraham, N.; et al. Helicobacter pylori -Negative Gastritis: Prevalence and Risk Factors. Am. J. Gastroenterol. 2013, 108, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Miftahussurur, M.; Waskito, L.A.; Syam, A.F.; Nusi, I.A.; Wibawa, I.D.N.; Rezkitha, Y.A.A.; Siregar, G.; Yulizal, O.; Akil, F.; Uwan, W.B.; et al. Analysis of risks of gastric cancer by gastric mucosa among Indonesian ethnic groups. PLoS ONE 2019, 14, e0216670. [Google Scholar] [CrossRef] [PubMed]
- Waskito, L.A.; Rezkitha, Y.A.A.; Vilaichone, R.-K.; Sugihartono, T.; Mustika, S.; Wibawa, I.D.N.; Yamaoka, Y.; Miftahussurur, M. The role of non-Helicobacter pylori bacteria in the pathogenesis of gastroduodenal diseases. Gut Pathog. 2022, 14, 19. [Google Scholar] [CrossRef]
- Miftahussurur, M.; Waskito, L.A.; El-Serag, H.B.; Ajami, N.J.; Nusi, I.A.; Syam, A.F.; Matsumoto, T.; Rezkitha, Y.A.A.; Doohan, D.; Fauzia, K.A.; et al. Gastric microbiota and Helicobacter pylori in Indonesian population. Helicobacter 2020, 25, e12695. [Google Scholar] [CrossRef]
- Gantuya, B.; El-Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Oyuntsetseg, K.; Azzaya, D.; Uchida, T.; Yamaoka, Y. Gastric Microbiota in Helicobacter pylori-Negative and -Positive Gastritis Among High Incidence of Gastric Cancer Area. Cancers 2019, 11, 504. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Cao, W.; Zhang, Z. Alterations of Gastric Microbiota in Gastric Cancer and Precancerous Stages. Front. Cell. Infect. Microbiol. 2021, 11, 559148. [Google Scholar] [CrossRef]
- Kim, H.-N.; Kim, M.-J.; Jacobs, J.P.; Yang, H.-J. Altered Gastric Microbiota and Inflammatory Cytokine Responses in Patients with Helicobacter pylori-Negative Gastric Cancer. Nutrients 2022, 14, 4981. [Google Scholar] [CrossRef]

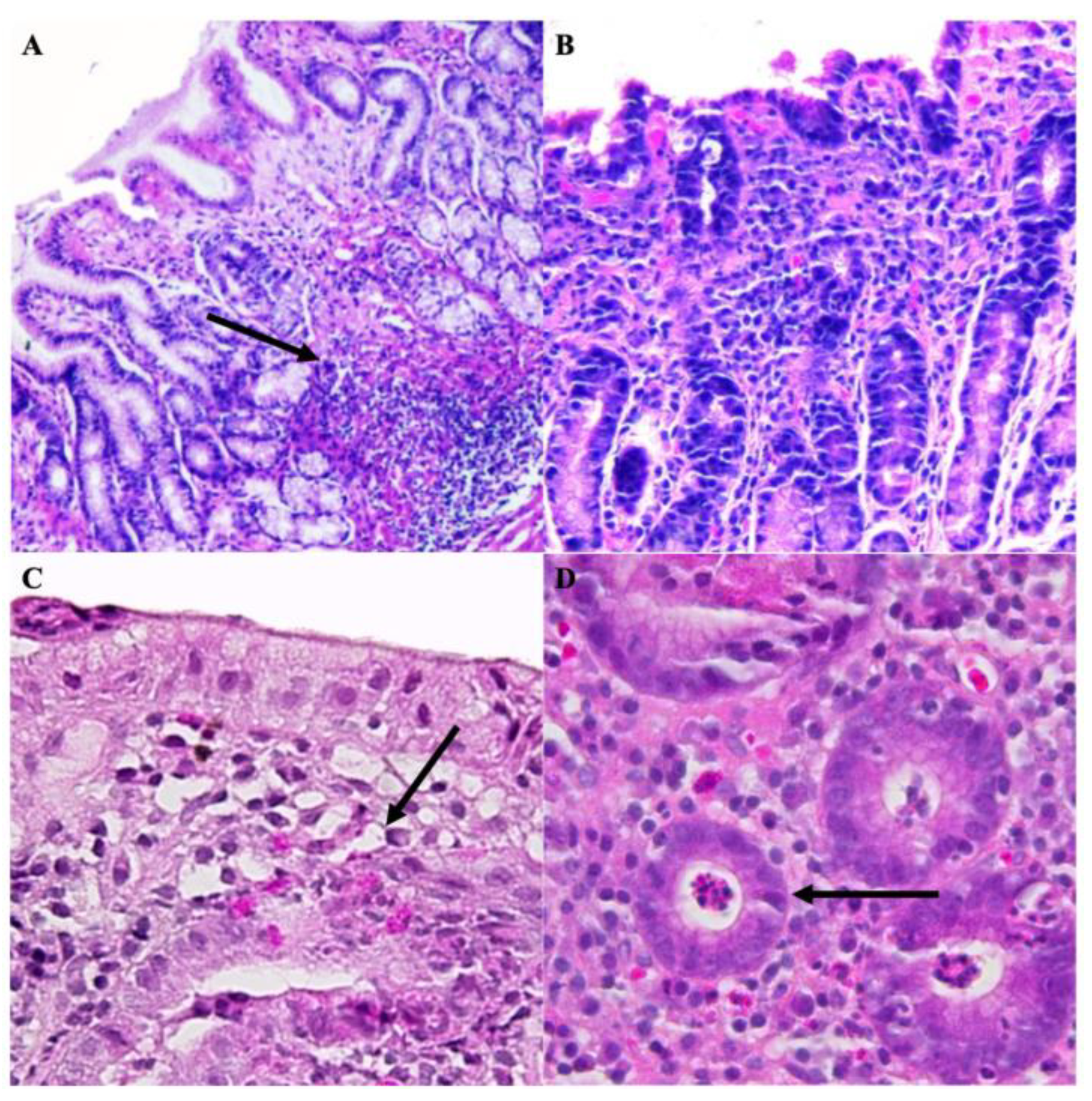
 ). Helicobacter representative sequences were observed in a posttreatment patient (highlighted with
). Helicobacter representative sequences were observed in a posttreatment patient (highlighted with  ), B: Pre-treatment patient. A: Post-treatment patient.
), B: Pre-treatment patient. A: Post-treatment patient.
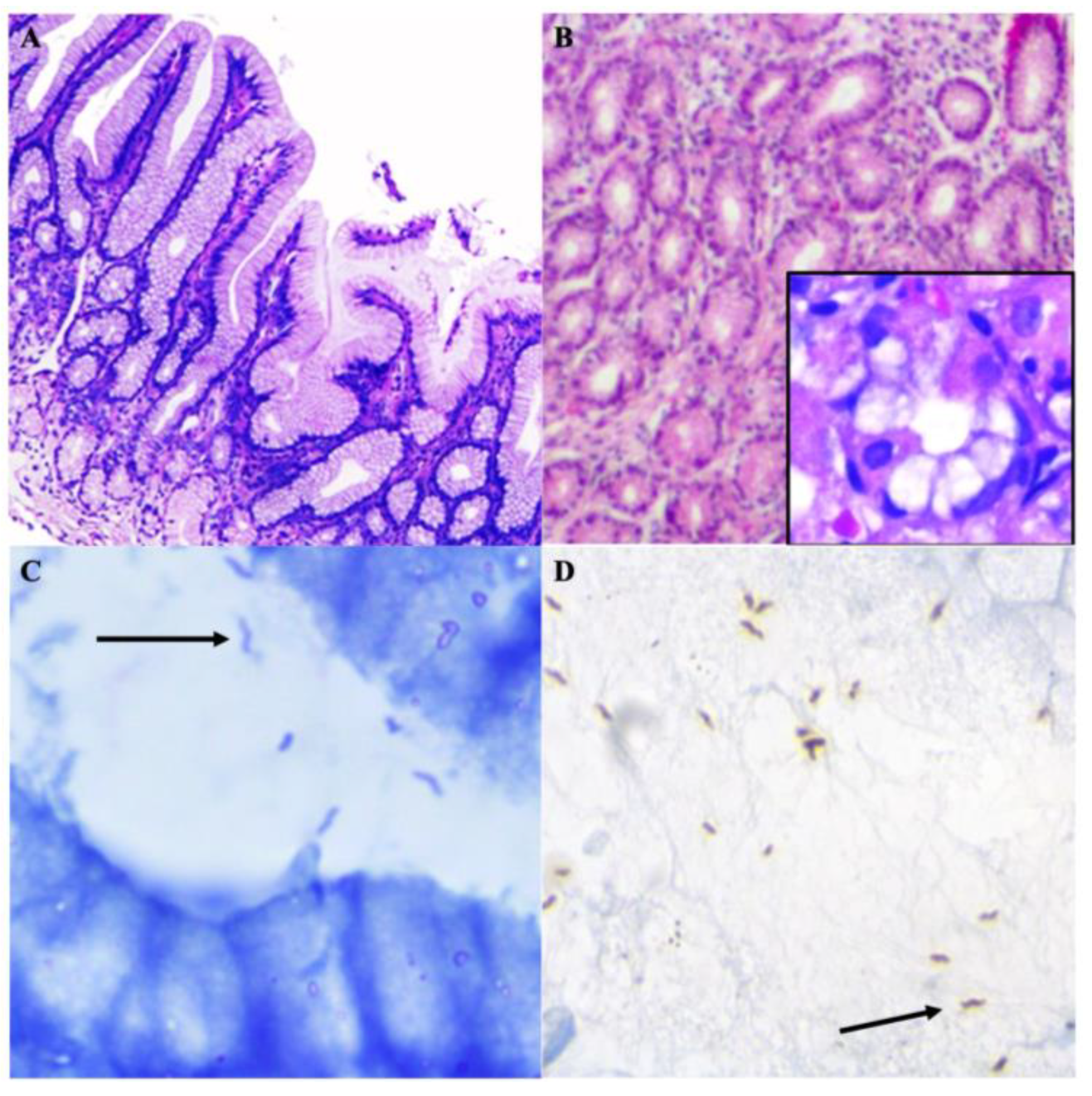
 ). Helicobacter representative sequences were observed in a posttreatment patient (highlighted with
). Helicobacter representative sequences were observed in a posttreatment patient (highlighted with  ), B: Pre-treatment patient. A: Post-treatment patient.
), B: Pre-treatment patient. A: Post-treatment patient.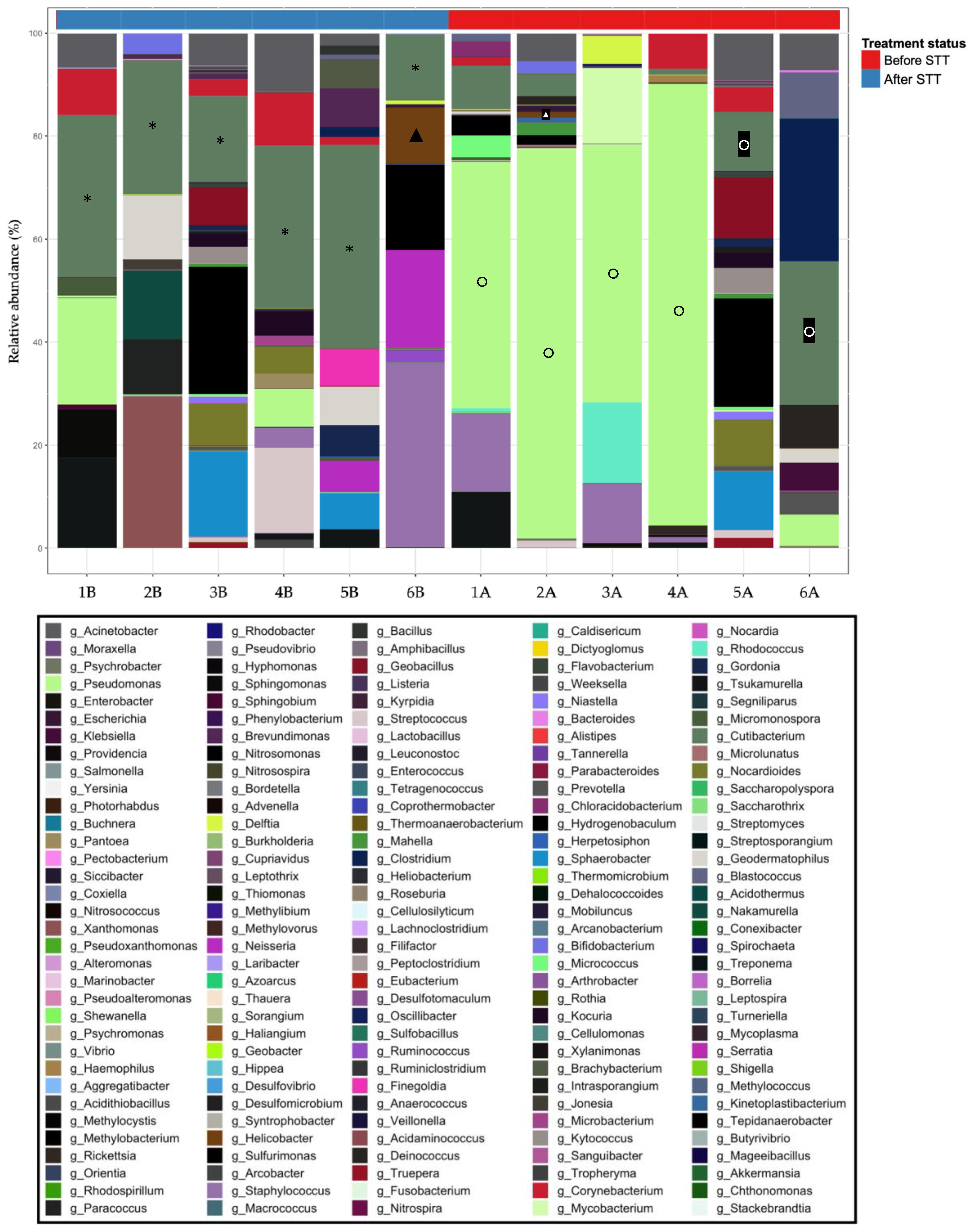

| Patients | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | 1B | 1A | 2B | 2A | 3B | 3A | 4B | 4A | 5B | 5A | 6B | 6A |
| Atrophy | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 |
| Mononuclear infiltrates | 2 | 1 | 0 | 0 | 1 | 1 | 3 | 2 | 2 | 3 | 1 | 1 |
| Neutrophilic infiltrates | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 1 | 0 | 1 |
| Metaplasia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Lymphoid follicles | 0 | 0 | 0 | 0 | 2 | 0 | 3 | 1 | 0 | 1 | 0 | 0 |
| Glandular atrophy | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| Foveolar hyperplasia | 2 60% | 0 | 0 | 0 | 0 | 0 | 2 50% | 2 60% | 0 | 1 30% | 1 30% | 1 30% |
| Regenerative changes | 1 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 0 |
| Bacillary structures | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 * | 0 | 0 |
| Patient | PCR | Urease | ||||
|---|---|---|---|---|---|---|
| glmM | cagA | vacA | ||||
| Before STT | After STT | Before STT | After STT | |||
| 1 | + *,c | − | + c | + c | + *,c | − |
| 2 | − | − | − | − | − | − |
| 3 | − | − | − | − | − | − |
| 4 | − | − | − | − | + *,c | − |
| 5 | + *,c | − | + c | + c | + *,c | − |
| 6 | + *,c | − | + c | + c | − | − |
| Patient | Good’s Coverage | Observed OTUs | ||
|---|---|---|---|---|
| B | A | B | A | |
| 1 | 1 | 1 | 27 | 27 |
| 2 | 1 | 1 | 34 | 40 |
| 3 | 1 | 1 | 42 | 29 |
| 4 | 1 | 1 | 35 | 37 |
| 5 | 1 | 1 | 51 | 44 |
| 6 | 1 | 1 | 31 | 43 |
| Patient | Shannon’s (H) | Faith’s PD | Chao1 | Simpson’s Index | ||||
|---|---|---|---|---|---|---|---|---|
| B | A | B | A | B | A | B | A | |
| 1 | 4.207 | 4.373 | 6.861 | 14.009 | 27 | 27 | 0.931 | 0.944 |
| 2 | 4.633 | 4.979 | 8.802 | 20.566 | 34 | 40 | 0.951 | 0.965 |
| 3 | 5.068 | 4.082 | 24.488 | 6.423 | 42 | 29 | 0.967 | 0.920 |
| 4 | 4.853 | 4.679 | 14.987 | 14.856 | 35 | 37 | 0.959 | 0.953 |
| 5 | 5.104 | 5.046 | 14.097 | 24.589 | 51 | 44 | 0.963 | 0.959 |
| 6 | 4.381 | 4.968 | 13.394 | 15.409 | 31 | 43 | 0.939 | 0.962 |
| Group 1 | Group 2 | n | Permutations | R | p | q |
|---|---|---|---|---|---|---|
| B | A | 12 | 999 | 0.073 | 0.273 | 0.273 |
| Good’s Coverage | Observed OTUs | Shannon’s (H) | Faith’s PD | Chao1 | Simpson’s Index | |
|---|---|---|---|---|---|---|
| Paired t test p-value | 1 | 1 | 0.914 | 0.643 | 1 | 0.933 |
| Wilcoxon (W) p-value | 1 | 0.81 | 0.749 | 0.423 | 0.81 | 0.936 |
| Metric | Pathobiont | |
|---|---|---|
| C. acnes | H. pylori | |
| Mean | 192,315,183.00 | 11,538.67 |
| Standard deviation | 203,649,007.30 | 3168.79 |
| n | 29 | 6 |
| Median | 157,942,606.40 | 11,973.67 |
| P25 | 36,511,873.51 | 9478.76 |
| P75 | 230,486,322.70 | 12,922.57 |
| Number of Transcripts (n = 7) | Median | 25th Percentile | 75th Percentile | Minimum | Maximum | p-Value |
|---|---|---|---|---|---|---|
| C. acnes | 16,915,313.11 | 8,383,738.74 | 36,511,872.51 | 0.90 | 48,151,777.41 | 0.017 |
| H. pylori | 11,383.78 | 6807.32 | 12,922.57 | 0.90 | 16,076.05 |
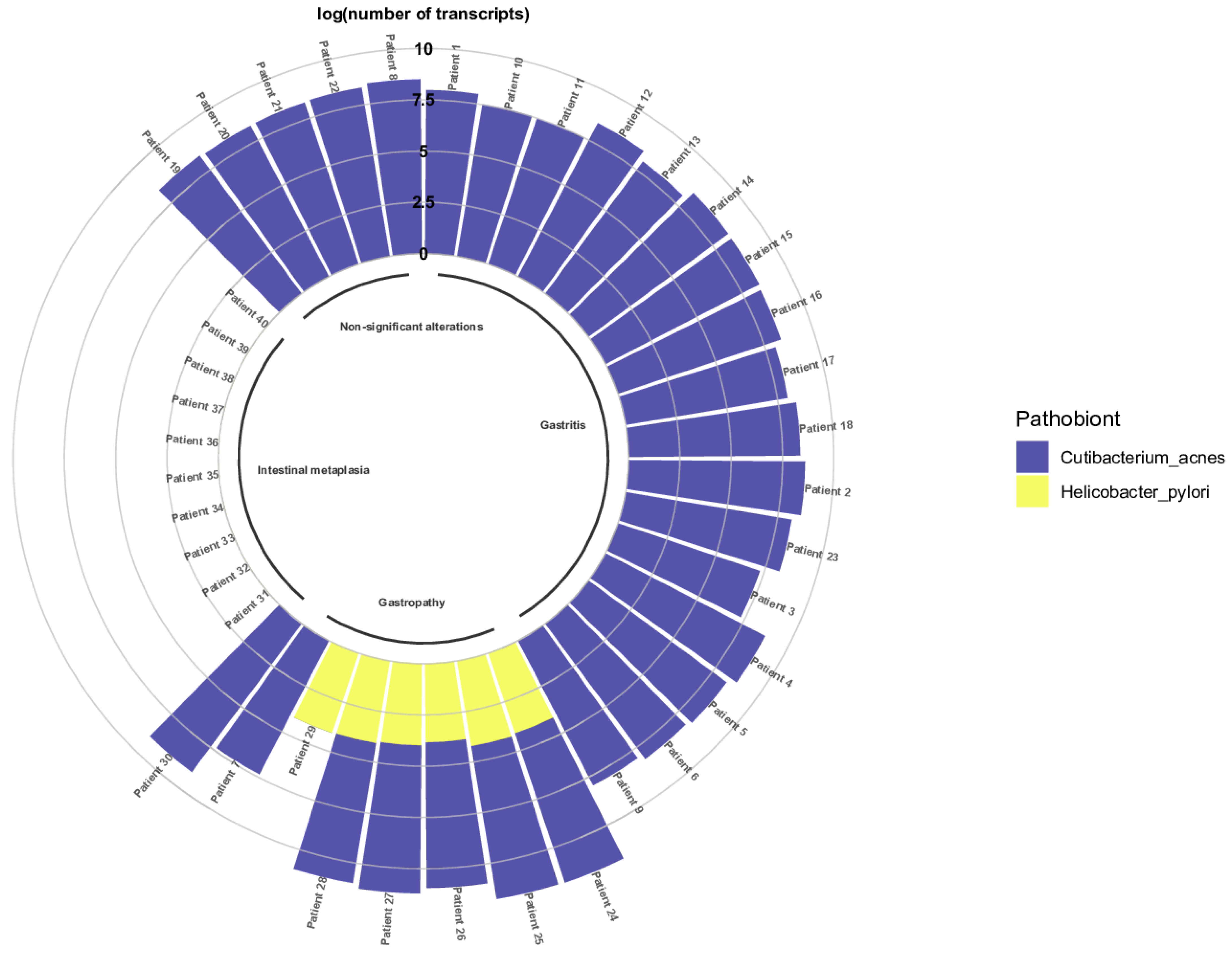
| Pathology | Median | 25th Percentile | 75th Percentile | Minimum | Maximum | p-Value |
|---|---|---|---|---|---|---|
| Gastritis | 157,942,606.40 | 94,564,890.45 | 272,850,376.48 | 19,229,694.53 | 573,263,008.44 | <0.001 |
| Gastropathy | 19,388,023.30 | 12,826,312.73 | 36,511,872.51 | 8,383,738.74 | 48,151,777.41 | |
| Non-significant alterations | 221,339,080.30 | 202,746,241.91 | 230,486,321.70 | 194,699,912.73 | 318,670,312.02 | |
| Intestinal metaplasia | <1 | <1 | <1 | <1 | 990,328,266.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez-Ramirez, U.; Nolasco-Romero, C.G.; Contreras-Rodríguez, A.; Zuñiga, G.; Mendoza-Elizalde, S.; Prado-Galbarro, F.-J.; Pérez Aguilar, F.; Pedraza Tinoco, J.E.; Valencia-Mayoral, P.; Velázquez-Guadarrama, N. Dysbiosis by Eradication of Helicobacter pylori Infection Associated with Follicular Gastropathy and Pangastropathy. Microorganisms 2023, 11, 2748. https://doi.org/10.3390/microorganisms11112748
Gomez-Ramirez U, Nolasco-Romero CG, Contreras-Rodríguez A, Zuñiga G, Mendoza-Elizalde S, Prado-Galbarro F-J, Pérez Aguilar F, Pedraza Tinoco JE, Valencia-Mayoral P, Velázquez-Guadarrama N. Dysbiosis by Eradication of Helicobacter pylori Infection Associated with Follicular Gastropathy and Pangastropathy. Microorganisms. 2023; 11(11):2748. https://doi.org/10.3390/microorganisms11112748
Chicago/Turabian StyleGomez-Ramirez, Uriel, Carolina G. Nolasco-Romero, Araceli Contreras-Rodríguez, Gerardo Zuñiga, Sandra Mendoza-Elizalde, Francisco-Javier Prado-Galbarro, Fernando Pérez Aguilar, Jonatan Elihu Pedraza Tinoco, Pedro Valencia-Mayoral, and Norma Velázquez-Guadarrama. 2023. "Dysbiosis by Eradication of Helicobacter pylori Infection Associated with Follicular Gastropathy and Pangastropathy" Microorganisms 11, no. 11: 2748. https://doi.org/10.3390/microorganisms11112748






