Natural Flavonoid Derivatives Have Pan-Coronavirus Antiviral Activity
Abstract
:1. Introduction
2. Methods
2.1. Study Design
2.2. Chemistry
2.3. In Silico Screening of Natural Compounds
2.4. SARS-CoV-2 3CLpro Assay
2.5. Cell Viability Assay to Determine Compounds’ Toxicity
2.6. SARS-CoV-2 In Vitro Assay
2.7. SARS-CoV-2 Titration
2.8. Feline-CoV, Bovine-CoV, and HCoV-OC43 In Vitro Assays and Titration
2.9. Coronaviruses In Vitro Assays
2.10. Statistical Analysis
3. Results
3.1. Virtual Screening of the in-House Natural Products Library
3.2. SARS-CoV-2 3CLpro Assay
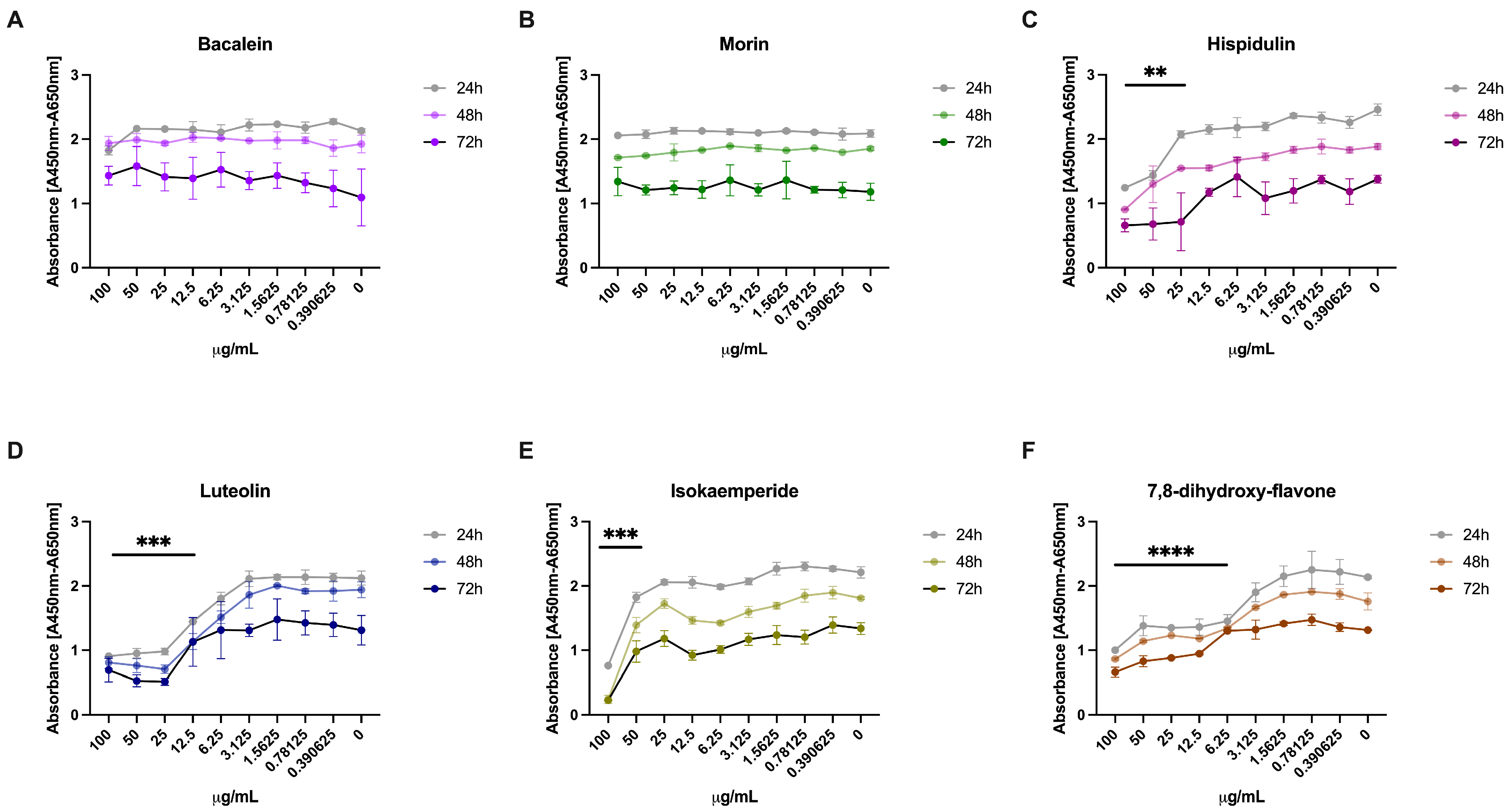
3.3. Effect of the 3CLpro Inhibitors on VERO E6 Viability
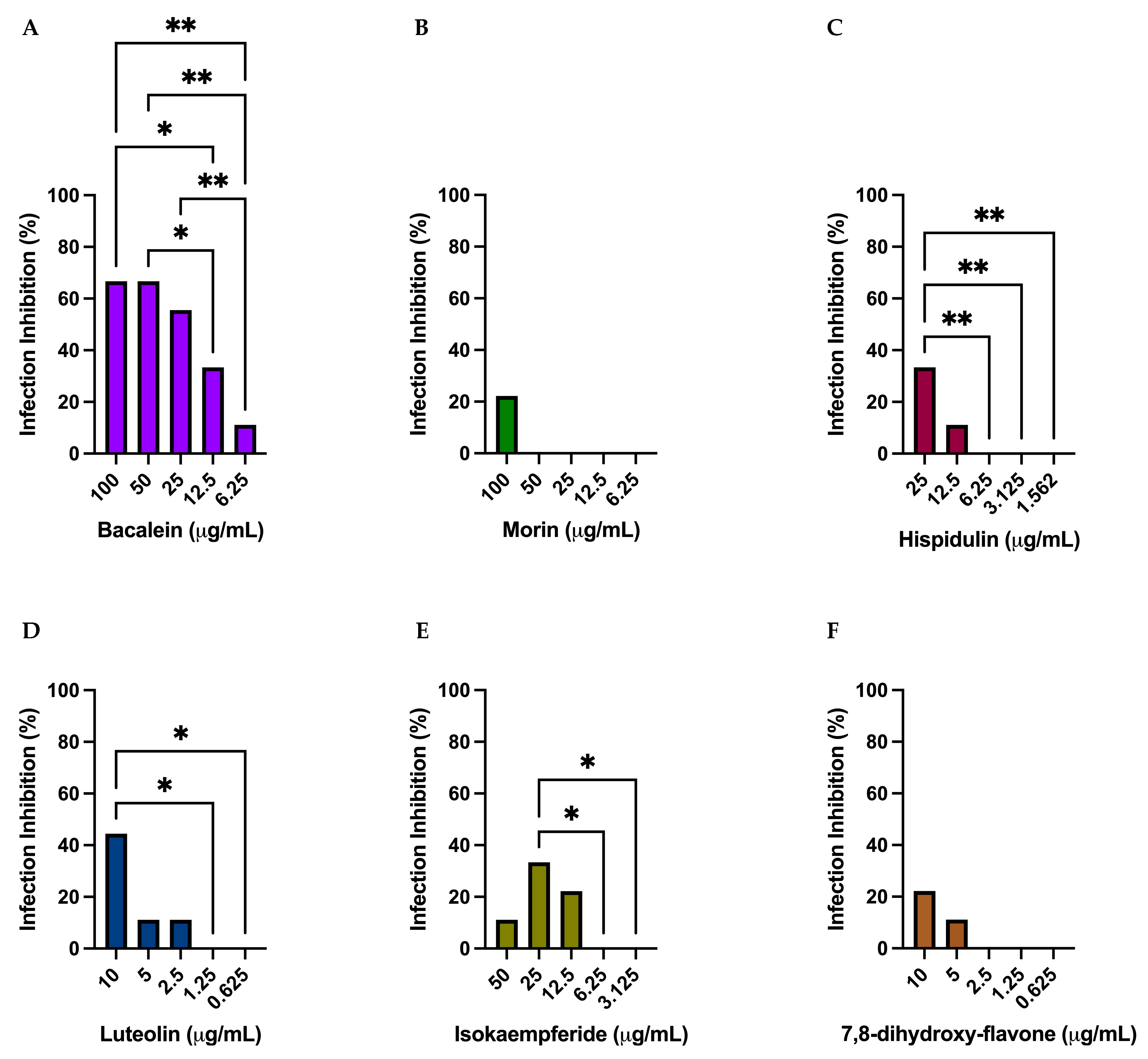
3.4. Antiviral Activity of 3CLpro Inhibitors against SARS-CoV-2
3.5. Effect of the 3CLpro Inhibitors on CRFK and HRT-18 Viability and Pan-Coronaviral Activity
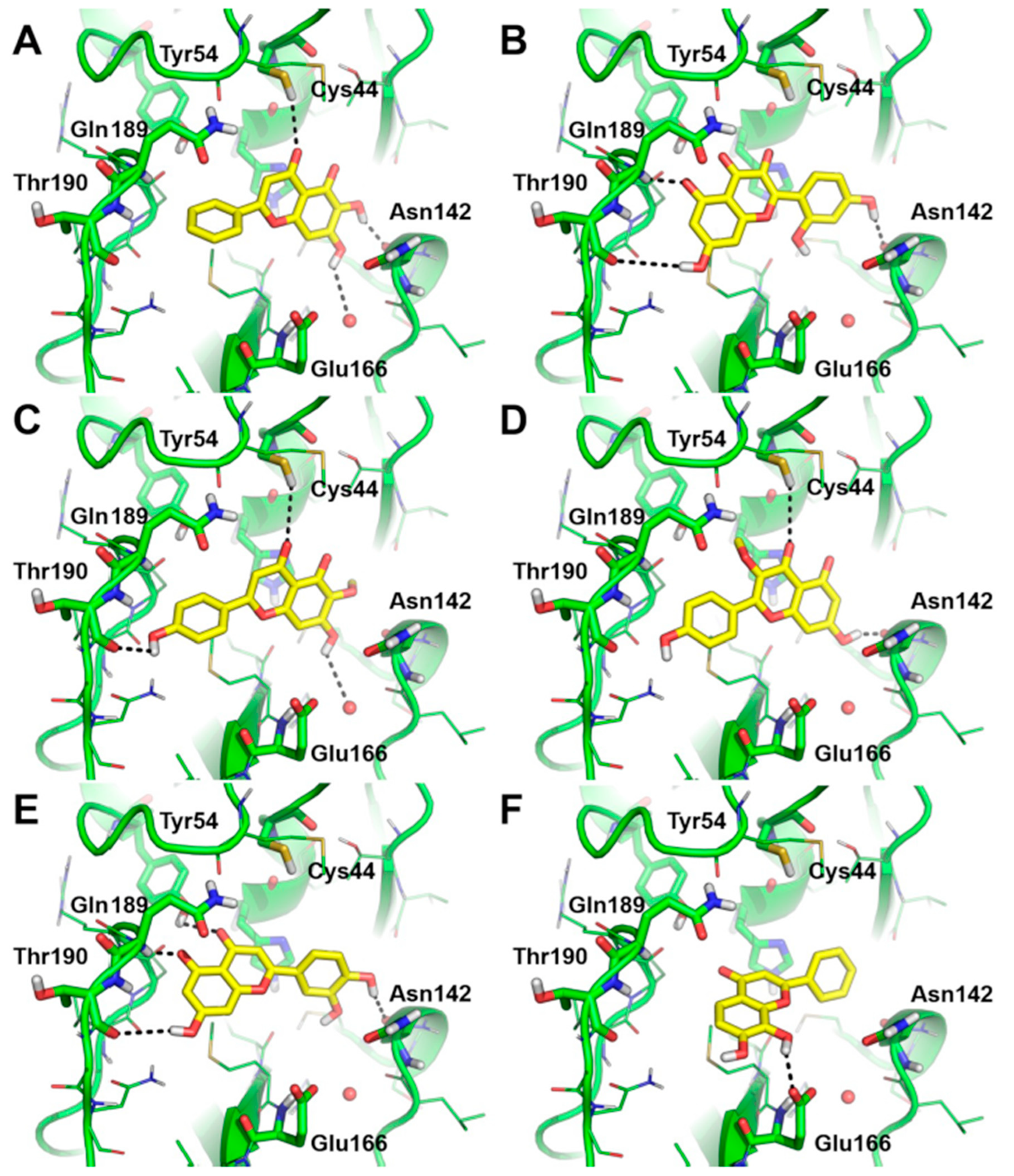
3.6. Predicted Binding Mode of Most Effective SARS-CoV-2 3CLpro Inhibitors
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hui, D.S.; Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.C.; Lu, C.C.; Bau, D.T.; Chiu, Y.J.; Yen, Y.T.; Hsu, Y.M.; Fu, C.W.; Kuo, S.C.; Lo, Y.S.; Chiu, H.Y.; et al. Approaches towards fighting the COVID-19 pandemic (Review). Int. J. Mol. Med. 2021, 47, 3–22. [Google Scholar] [CrossRef]
- Jo, S.; Kim, S.; Kim, D.Y.; Kim, M.S.; Shin, D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhib. Med. Chem. 2020, 35, 1539–1544. [Google Scholar] [CrossRef]
- Rehman, M.T.; AlAjmi, M.F.; Hussain, A. Natural Compounds as Inhibitors of SARS-CoV-2 Main Protease (3CLpro): A Molecular Docking and Simulation Approach to Combat COVID-19. Curr Pharm Des. 2021, 27, 3577–3589. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Remuzzi, G.; Schiaffino, S.; Santoro, M.G.; FitzGerald, G.A.; Melino, G.; Patrono, C. Drugs for the prevention and treatment of COVID-19 and its complications: An update on what we learned in the past 2 years. Front. Pharmacol. 2020, 13, 987816. [Google Scholar] [CrossRef]
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of COVID-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Akinosoglou, K.; Schinas, G.; Gogos, C. Oral Antiviral Treatment for COVID-19: A Comprehensive Review on Nirmatrelvir/Ritonavir. Viruses 2022, 14, 2540. [Google Scholar] [CrossRef]
- Vangeel, L.; Chiu, W.; De Jonghe, S.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Leyssen, P.; Neyts, J.; Jochmans, D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022, 198, 105252. [Google Scholar] [CrossRef] [PubMed]
- Rubin, R. From Positive to Negative to Positive Again—The Mystery of Why COVID-19 Rebounds in Some Patients Who Take Paxlovid. JAMA 2022, 327, 2380–2382. [Google Scholar] [CrossRef]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef]
- Clementi, N.; Scagnolari, C.; D’Amore, A.; Palombi, F.; Criscuolo, E.; Frasca, F.; Pierangeli, A.; Mancini, N.; Antonelli, G.; Clementi, M.; et al. Naringenin is a powerful inhibitor of SARS-CoV-2 infection in vitro. Pharmacol. Res. 2021, 163, 105255. [Google Scholar] [CrossRef]
- Kaul, R.; Paul, P.; Kumar, S.; Büsselberg, D.; Dwivedi, V.D.; Chaari, A. Promising Antiviral Activities of Natural Flavonoids against SARS-CoV-2 Targets: Systematic Review. Int. J. Mol. Sci. 2021, 22, 11069. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef]
- Theoharides, T.C.; Cholevas, C.; Polyzoidis, K.; Politis, A. Long-COVID syndrome-associated brain fog and chemofog: Luteolin to the rescue. Biofactors 2021, 47, 232–241. [Google Scholar] [CrossRef]
- Bardelčíková, A.; Miroššay, A.; Šoltýs, J.; Mojžiš, J. Therapeutic and prophylactic effect of flavonoids in post-COVID-19 therapy. Phytother. Res. 2022, 36, 2042–2060. [Google Scholar] [CrossRef]
- Jo, S.; Kim, H.; Kim, S.; Shin, D.H.; Kim, M.S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019, 94, 2023–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theerawatanasirikul, S.; Thangthamniyom, N.; Kuo, C.J.; Semkum, P.; Phecharat, N.; Chankeeree, P.; Lekcharoensuk, P. Natural Phytochemicals, Luteolin and Isoginkgetin, Inhibit 3C Protease and Infection of FMDV, In Silico and In Vitro. Viruses 2021, 13, 2118. [Google Scholar] [CrossRef] [PubMed]
- Ghirga, F.; Quaglio, D.; Mori, M.; Cammarone, S.; Iazzetti, A.; Goggiamani, A.; Ingallina, C.; Botta, B.; Calcaterra, A. A unique high-diversity natural product collection as a reservoir of new therapeutic leads. Org. Chem. Front. 2021, 8, 996–1025. [Google Scholar] [CrossRef]
- Angiosperm Phylogeny, G.; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Wiczkowski, W.; Romaszko, J.; Bucinski, A.; Szawara-Nowak, D.; Honke, J.; Zielinski, H.; Piskula, M.K. Quercetin from shallots (Allium cepa L. var. aggregatum) is more bioavailable than its glucosides. J. Nutr. 2008, 138, 885–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Häkkinen, S.H.; Kärenlampi, S.O.; Heinonen, I.M.; Mykkänen, H.M.; Törrönen, A.R. Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J. Agric. Food Chem. 1999, 47, 2274–2279. [Google Scholar] [CrossRef]
- Sakawa, Y. Chemical constituents of Alnus sieboldiana (Betulaceae) II. The isolation and structure of flavonoids and stilbenes. BCSJ 1971, 44, 2761–2766. [Google Scholar]
- Moghaddam, G.; Ebrahimi, S.A.; Rahbar-Roshandel, N.; Foroumadi, A. Antiproliferative activity of flavonoids: Influence of the sequential methoxylation state of the flavonoid structure. Phytother. Res. 2012, 26, 1023–1028. [Google Scholar] [CrossRef]
- Bradburn, M.J.; Clark, T.G.; Love, S.B.; Altman, D.G. Survival analysis Part III: Multivariate data analysis–choosing a model and assessing its adequacy and fit. Br. J. Cancer 2003, 89, 605–611. [Google Scholar] [CrossRef]
- Arbos, P.; Arangoa, M.A.; Campanero, M.A.; Irache, J.M. Quantification of the bioadhesive properties of protein-coated PVM/MA nanoparticles. Int. J. Pharm. 2002, 242, 129–136. [Google Scholar] [CrossRef]
- Kaur, A.; Singh, R.; Dey, C.S.; Sharma, S.S.; Bhutani, K.K.; Singh, I.P. Antileishmanial phenylpropanoids from Alpinia galanga (Linn.) Willd. Indian J. Exp. Biol. 2010, 48, 314–317. [Google Scholar]
- Sotnikova, O.M.; Chagovets, R.K.; Litvinenko, V.I. New flavanone compounds from Euphorbia stepposa. Chem. Nat. Compd. 1968, 4, 71–74. [Google Scholar] [CrossRef]
- Beutler, J.A.; Cardellina Ii, J.H.; Lin, C.M.; Hamel, E.; Cragg, G.M.; Boyd, M.R. Centaureidin, a cytotoxic flavone from Polymnia fruticosa, inhibits tubulin polymerization. Bioorg. Med. Chem. Lett. 1993, 3, 581–584. [Google Scholar] [CrossRef]
- Wagner, H.; Hörhammer, L.; Aurnhammer, G.; Farkas, L. Strukturaufklärung und Synthese des Didymins, eines Isosakuranetin–7-β-rutinosids aus Monarda didyma L. Chem. Ber. 1968, 101, 445–449. [Google Scholar] [CrossRef]
- Sowndhararajan, K.; Deepa, P.; Kim, M.; Park, S.J.; Kim, S. Baicalein as a potent neuroprotective agent: A review. Biomed. Pharmacother. 2017, 95, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.X.; Xu, Y.J.; Zhao, D.X.; Ma, F.S. A comparison between hairy root cultures and wild plants of Saussurea involucrata in phenylpropanoids production. Plant Cell Rep. 2006, 24, 750–754. [Google Scholar] [CrossRef]
- Siraichi, J.T.G.; Felipe, D.F.; Brambilla, L.Z.S.; Gatto, M.J.; Terra, V.A.; Cecchini, A.L.; Cortez, L.E.R.; Rodrigues-Filho, E.; Cortez, D.A.G. Antioxidant capacity of the leaf extract obtained from Arrabidaea chica cultivated in Southern Brazil. PLoS ONE 2013, 8, e72733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, R.; Natesan, V. Chrysin: Sources, beneficial pharmacological activities, and molecular mechanism of action. Phytochemistry 2018, 145, 187–196. [Google Scholar] [CrossRef]
- Asmi, K.S.; Lakshmi, T.; Balusamy, S.R.; Parameswari, R. Therapeutic aspects of taxifolin—An update. J. Adv. Pharm. Res. 2017, 7, 187–189. [Google Scholar]
- Halevas, E.; Mavroidi, B.; Kaplanis, M.; Hatzidimitriou, A.G.; Moschona, A.; Litsardakis, G.; Pelecanou, M. Hydrophilic bis-MPA hyperbranched dendritic scaffolds as nanocarriers of a fully characterized flavonoid morin-Zn (II) complex for anticancer applications. J. Inorg. Biochem. 2022, 232, 111832. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Kang, K.; Jho, E.H.; Jung, Y.J.; Nho, C.W.; Um, B.H.; Pan, C.H. Hepatoprotective effect of flavonoid glycosides from Lespedeza cuneata against oxidative stress induced by tert-butyl hyperoxide. Phytother. Res. 2011, 25, 1011–1017. [Google Scholar] [CrossRef]
- Wollenweber, E.; Mann, K.; Arriaga-Giner, F.J.; Roitman, J.N.; West, J.G. Exudate flavonoids from two Australian Asteraceae, Bracteantha viscosa and Cassinia quinquefaria. Phytochemistry 1993, 33, 871–873. [Google Scholar] [CrossRef]
- Yang, H.; Gan, C.; Guo, Y.; Qu, L.; Ma, S.; Ren, Y.; Wang, X.; Wang, L.; Huang, J.; Wang, J. Two novel compounds from green walnut husks (Juglans mandshurica Maxim.). Nat. Prod. Res. 2022, 36, 3389–3395. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Li, Y.; Lin, L.; Sayasith, P.R.; Tarr, A.T.; Wright, E.B.; Yasmin, S.; Lannigan, D.A.; O’Doherty, G.A. Synthesis and Biological Evaluation of 4′-Substituted Kaempfer-3-ols. J. Org. Chem. 2020, 85, 4279–4288. [Google Scholar] [CrossRef]
- Li, H.X.; Park, J.U.; Su, X.D.; Kim, K.T.; Kang, J.S.; Kim, Y.R.; Kim, Y.H.; Yang, S.Y. Identification of anti-melanogenesis constituents from Morus alba L. leaves. Molecules 2018, 23, 2559. [Google Scholar] [CrossRef] [Green Version]
- Kirubakaran, P.; Muthusamy, K.; Dhanachandra Singh, K.; Nagamani, S. Homology modeling, molecular dynamics, and molecular docking studies of Trichomonas vaginalis carbamate kinase. Med. Chem. Res. 2012, 21, 2105–2116. [Google Scholar] [CrossRef]
- Zahran, E.M.; Abdelmohsen, U.R.; Hussein, A.S.; Salem, M.A.; Khalil, H.E.; Yehia Desoukey, S.; Fouad, M.A.; Kamel, M.S. Antiulcer potential and molecular docking of flavonoids from Ocimum forskolei Benth. family Lamiaceae. Nat. Prod. Res. 2021, 35, 1933–1937. [Google Scholar] [CrossRef]
- Meng, Q.; Li, G.; Luo, B.; Wang, L.; Lu, Y.; Liu, W. Screening and isolation of natural antioxidants from Ziziphora clinopodioides Lam. with high performance liquid chromatography coupled to a post-column Ce (IV) reduction capacity assay. RSC Adv. 2016, 6, 62378–62384. [Google Scholar] [CrossRef]
- Chen, L.C.; Hsu, K.C.; Chiou, L.C.; Tseng, H.J.; Huang, W.J. Total synthesis and metabolic stability of hispidulin and its d-labelled derivative. Molecules 2017, 22, 1897. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Kaur, M.; Vyas, B.; Silakari, O. Design, synthesis and biological evaluation of 2-Phenyl-4H-chromen-4-one derivatives as polyfunctional compounds against Alzheimer’s disease. Med. Chem. Res. 2018, 27, 520–530. [Google Scholar] [CrossRef]
- Xu, S.; Shang, M.Y.; Liu, G.X.; Xu, F.; Wang, X.; Shou, C.C.; Cai, S.Q. Chemical constituents from the rhizomes of Smilax glabra and their antimicrobial activity. Molecules 2013, 18, 5265–5287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douangamath, A.; Fearon, D.; Gehrtz, P.; Krojer, T.; Lukacik, P.; Owen, C.D.; Resnick, E.; Strain-Damerell, C.; Aimon, A.; Abranyi-Balog, P.; et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef] [PubMed]
- Picarazzi, F.; Zuanon, M.; Pasqualetto, G.; Cammarone, S.; Romeo, I.; Young, M.T.; Brancale, A.; Bassetto, M.; Mori, M. Identification of Small Molecular Chaperones Binding P23H Mutant Opsin through an In Silico Structure-Based Approach. J. Chem. Inf. Model 2022, 62, 5794–5805. [Google Scholar] [CrossRef]
- Platella, C.; Ghirga, F.; Zizza, P.; Pompili, L.; Marzano, S.; Pagano, B.; Quaglio, D.; Vergine, V.; Cammarone, S.; Botta, B.; et al. Identification of Effective Anticancer G-Quadruplex-Targeting Chemotypes through the Exploration of a High Diversity Library of Natural Compounds. Pharmaceutics 2020, 13, 1611. [Google Scholar] [CrossRef]
- Kuchlyan, J.; Martinez-Fernandez, L.; Mori, M.; Gavvala, K.; Ciaco, S.; Boudier, C.; Richert, L.; Didier, P.; Tor, Y.; Improta, R.; et al. What Makes Thienoguanosine an Outstanding Fluorescent DNA Probe? J. Am. Chem. Soc. 2020, 142, 16999–17014. [Google Scholar] [CrossRef] [PubMed]
- OEDOCKING 3.3.0.3: OpenEye Scientific Software, Inc. Santa Fe, NM. Available online: http://www.eyesopen.com (accessed on 27 December 2022).
- McGann, M. FRED Pose Prediction and Virtual Screening Accuracy. J. Chem. Inf. Model. 2011, 51, 578–596. [Google Scholar] [CrossRef]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and the Cambridge Structural Database. J. Chem. Inf. Model 2010, 50, 572–584. [Google Scholar] [CrossRef]
- QUACPAC 2.0.0.3: OpenEye Scientific Software, Santa Fe, NM. Available online: http://www.eyesopen.com (accessed on 27 December 2022).
- SZYBKI 1.10.0.3: OpenEye Scientific Software, Santa Fe, NM. Available online: http://www.eyesopen.com (accessed on 27 December 2022).
- Criscuolo, E.; Giuliani, B.; Ferrari, D.; Ferrarese, R.; Diotti, R.A.; Clementi, M.; Mancini, N.; Clementi, N. Proper Selection of In Vitro Cell Model Affects the Characterization of the Neutralizing Antibody Response against SARS-CoV-2. Viruses 2022, 14, 1232. [Google Scholar] [CrossRef]
- Romeo, A.; Iacovelli, F.; Scagnolari, C.; Scordio, M.; Frasca, F.; Condò, R.; Ammendola, S.; Gaziano, R.; Anselmi, M.; Divizia, M.; et al. Potential Use of Tea Tree Oil as a Disinfectant Agent against Coronaviruses: A Combined Experimental and Simulation Study. Molecules 2022, 27, 3786. [Google Scholar] [CrossRef]
- Kashyap, P.; Thakur, M.; Singh, N.; Shikha, D.; Kumar, S.; Baniwal, P.; Yadav, Y.S.; Sharma, M.; Sridhar, K.; Inbaraj, B.S. In Silico Evaluation of Natural Flavonoids as a Potential Inhibitor of Coronavirus Disease. Molecules 2022, 27, 6374. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.H. An overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Scholle, F.; Kisthardt, S.C.; Xie, D.Y. Flavonols and dihydroflavonols inhibit the main protease activity of SARS-CoV-2 and the replication of human coronavirus 229E. Virology 2022, 571, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, H.H.; Shin, Y.S.; Kang, H.; Cho, H. The anti-HSV-1 effect of quercetin is dependent on the suppression of TLR-3 in Raw 264.7 cells. Arch. Pharm. Res. 2017, 40, 623–630. [Google Scholar] [CrossRef]
- Ganesan, S.; Faris, A.N.; Comstock, A.T.; Wang, Q.; Nanua, S.; Hershenson, M.B.; Sajjan, U.S. Quercetin inhibits rhinovirus replication in vitro and in vivo. Antivir. Res. 2012, 94, 258–271. [Google Scholar] [CrossRef]
- Lopes, B.R.P.; da Costa, M.F.; Genova Ribeiro, A.; da Silva, T.F.; Lima, C.S.; Caruso, I.P.; de Araujo, G.C.; Kubo, L.H.; Iacovelli, F.; Falconi, M.; et al. Quercetin pentaacetate inhibits in vitro human respiratory syncytial virus adhesion. Virus Res. 2020, 276, 197805. [Google Scholar] [CrossRef]
- Mehrbod, P.; Abdalla, M.A.; Fotouhi, F.; Heidarzadeh, M.; Aro, A.O.; Eloff, J.N.; McGaw, L.J.; Fasina, F.O. Immunomodulatory properties of quercetin-3-O-α-L-rhamnopyranoside from Rapanea melanophloeos against influenza a virus. BMC Complement. Altern. Med. 2018, 18, 184. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Zhu, G.H.; Zhang, Y.N.; Hu, Q.; Wang, H.N.; Yu, H.N.; Qin, X.Y.; Guan, X.Q.; Xiang, Y.W.; Tang, H.; et al. Flavonoids in Ampelopsis grossedentata as covalent inhibitors of SARS-CoV-2 3CLpro: Inhibition potentials, covalent binding sites and inhibitory mechanisms. Int J. Biol Macromol. 2021, 187, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Gravina, H.D.; Tafuri, N.F.; Silva Júnior, A.; Fietto, J.L.; Oliveira, T.T.; Diaz, M.A.; Almeida, M.R. In vitro assessment of the antiviral potential of trans-cinnamic acid, quercetin and morin against equid herpesvirus 1. Res. Vet. Sci. 2011, 91, e158–e162. [Google Scholar] [CrossRef]
- Bang, S.; Quy Ha, T.K.; Lee, C.; Li, W.; Oh, W.K.; Shim, S.H. Antiviral activities of compounds from aerial parts of Salvia plebeia R. Br. J. Ethnopharmacol. 2016, 192, 398–405. [Google Scholar] [CrossRef]
- Liu, H.; Ye, F.; Sun, Q.; Liang, H.; Li, C.; Li, S.; Lu, R.; Huang, B.; Tan, W.; Lai, L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J. Enzyme Inhib. Med. Chem. 2021, 36, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Onyango, H.; Odhiambo, P.; Angwenyi, D.; Okoth, P. In Silico Identification of New Anti-SARS-CoV-2 Main Protease (Mpro) Molecules with Pharmacokinetic Properties from Natural Sources Using Molecular Dynamics (MD) Simulations and Hierarchical Virtual Screening. J. Trop Med. 2022, 2022, 3697498. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, D.; Zhang, J.; Yin, X.; Li, J. Crystal structure of SARS-CoV 3C-like protease with baicalein. Biochem. Biophys. Res. Commun. 2022, 611, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Hong, S.J.; Sheng, J.; Bahari, F.; Culbertson, B.; Atanaki, F.F.; Aditham, A.K.; Kratz, A.F.; Luck, M.I.; Tian, R.; et al. Functional map of SARS-CoV-2 3CL protease reveals tolerant and immutable sites. Cell Host Microbe 2022, 30, 1354–1362. [Google Scholar] [CrossRef]
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Chand, M.A.; Brown, C.S.; Aarons, E.; Tong, C.; Langrish, C.; Hoschler, K.; Brown, K.; Galiano, M.; Myers, R.; et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill. 2012, 17, 20290. [Google Scholar] [CrossRef]
- Ciotti, M.; Angeletti, S.; Minieri, M.; Giovannetti, M.; Benvenuto, D.; Pascarella, S.; Sagnelli, C.; Bianchi, M.; Bernardini, S.; Ciccozzi, M. COVID-19 Outbreak: An Overview. Chemotherapy 2019, 64, 215–223. [Google Scholar] [CrossRef]
- Camero, M.; Lanave, G.; Catella, C.; Lucente, M.S.; Decaro, N.; Martella, V.; Buonavoglia, C. Evaluation of virucidal activity of fabrics using feline coronavirus. J. Virol. Methods 2021, 295, 114214. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.A.; et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef] [PubMed]
- Bhavanam, S.; Lee, B.; Qiu, Y.; Zelyas, N.; Pang, X.L. Evaluation of compressed sodium chloride on the inactivation of SARS-CoV-2 and surrogates. PLoS ONE 2022, 17, e0277881. [Google Scholar] [CrossRef]
- Gorse, G.J.; Donovan, M.M.; Patel, G.B.; Balasubramanian, S.; Lusk, R.H. Coronavirus and Other Respiratory Illnesses Comparing Older with Young Adults. Am. J. Med. 2015, 128, 11–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebihara, T.; Endo, R.; Ma, X.; Ishiguro, N.; Kikuta, H. Detection of human coronavirus NL63 in young children with bronchiolitis. J. Med. Virol. 2005, 75, 463–465. [Google Scholar] [CrossRef]
- Tsou, P.; Vadivelan, A.; Kovvuri, M.; Garg, N.; Thangavelu, M.; Wang, Y.; Raj, S. Association between multiple respiratory viral infections and pediatric intensive care unit admission among infants with bronchiolitis. Arch. Pediatr. 2020, 27, 39–44. [Google Scholar] [CrossRef]
- Al-Khayri, J.M.; Sahana, G.R.; Nagella, P.; Joseph, B.V.; Alessa, F.M.; Al-Mssallem, M.Q. Flavonoids as Potential Anti-Inflammatory Molecules: A Review. Molecules 2022, 27, 2901. [Google Scholar] [CrossRef]
- Saeedi-Boroujeni, A.; Mahmoudian-Sani, M.R. Anti-inflammatory potential of Quercetin in COVID-19 treatment. J. Inflamm. 2021, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Rui, W.; Li, S.; Xiao, H.; Xiao, M.; Shi, J. Baicalein Attenuates Neuroinflammation by Inhibiting NLRP3/caspase-1/GSDMD Pathway in MPTP Induced Mice Model of Parkinson’s Disease. Int. J. Neuropsychopharmacol. 2020, 23, 762–773. [Google Scholar] [CrossRef]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T.; et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef]
- Dai, C.; Li, H.; Wang, Y.; Tang, S.; Velkov, T.; Shen, J. Inhibition of Oxidative Stress and ALOX12 and NF-κB Pathways Contribute to the Protective Effect of Baicalein on Carbon Tetrachloride-Induced Acute Liver Injury. Antioxidants 2021, 10, 976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.; Shi, W.; Zhang, W.; Zhu, Y.; Zhang, Y.; Xie, Q.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, C.E.; Yount, B.L.; Graham, R.L.; Leist, S.R.; Hou, Y.J.; Dinnon, K.H., 3rd; Sims, A.C.; Swanstrom, J.; Gully, K.; Scobey, T.D.; et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA 2020, 117, 26915–26925. [Google Scholar] [CrossRef]
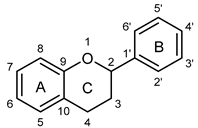 Basic Flavonoid Skeleton | ||||||
| Mol. | Common Name | Chemical Structure | M.W. | Molecular Formula | Source | Reference |
| Flavanones | ||||||
| 1 | Morin | 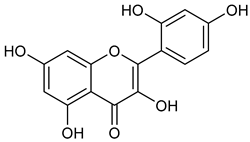 | 302.24 | C15H10O7 | Moriaceae family | [22] |
| 2 | Quercetin |  | 302.24 | C15H10O7 | Ginkgo biloba (Ginkgoaceae family) Hypericum perforatum (Hypericaceae family) Sambucus canadensis (Adoxaceae family) | [23,24] |
| 3 | Alnusin | 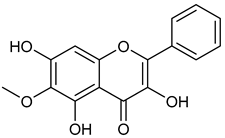 | 300.27 | C16H12O6 | Xerochrysum viscosum (Asteraceae family) Alnus sieboldiana (Betulaceae family) | [25] |
| 4 | Isokaempferide | 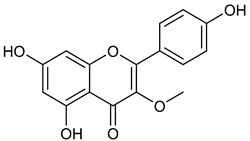 | 300.27 | C16H12O6 | Amburana cearensis (Fabacee family) | [26] |
| 5 | Galangin |  | 270.24 | C15H10O5 | Alpinia officinarum (Zingiberaceae family) Helichrysum aureonitens (Asteraceae family) Alpinia galanga (Zingiberaceae family) | [27,28,29] |
| Flavanones | ||||||
| 6 | Steppogenin |  | 288.26 | C15H12O6 | Euphorbia nicaeensis (Euforbiacee family) Maclura tricuspidata (Moraceae family) | [30] |
| 7 | Sakuranetin | 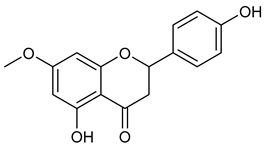 | 286.28 | C16H14O5 | Polymnia fruticosa (Araliaceae family) | [31] |
| 8 | Isosakuranetin | 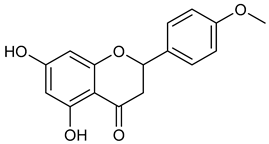 | 286.28 | C16H14O5 | Monarda didyma (Lamiaceae family) | [32] |
| Flavones | ||||||
| 9 | Baicalein | 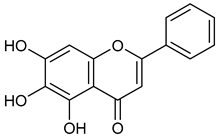 | 270.24 | C15H10O5 | Scutellaria baicalensis (Lamiaceae family) | [33] |
| 10 | Hispidulin | 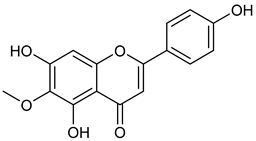 | 300.27 | C16H12O6 | Crossostephium chinense, Grindelia argentina and Saussurea involucrate (Asteraceae family) Arrabidaea chica (Bignoniaceae Family) | [34,35] |
| 11 | Chrysin |  | 254.24 | C15H10O4 | Passiflora caerulea and Passiflora incarnata (Passifloraceae family) Oroxylum indicum (Bignoniaceae family) | [36] |
| Flavanonol | ||||||
| 12 | Taxifolin | 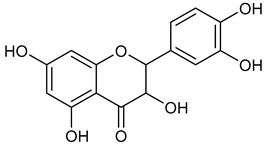 | 304.25 | C15H12O7 | Pinus roxburghii, Cedrus deodara (Pinaceae family) | [37] |
| Compound | Molecular Model | Concentrations Tested (µg/mL) | Percentage of Inhibition of 3CLpro |
|---|---|---|---|
| Morin | Docking | 100.00 | 53.64 (33.85) |
| 50.00 | 18.64 (4.36) | ||
| 25.00 | <10 | ||
| 12.5 | <10 | ||
| Baicalein | Docking | 100.00 | 94.72 (9.11) |
| 50.00 | 72.14 (7.75) | ||
| 25.00 | 25.57 (6.67) | ||
| 12.5 | 17.28 (1.11) | ||
| Hispidulin | Docking | 100.00 | 55.62 (17.04) |
| 50.00 | 29.76 (12.56) | ||
| 25.00 | <10 | ||
| 12.5 | <10 | ||
| Luteolin | Docking | 100.00 | 38.12 (2.32) |
| 50.00 | 30.88 (3.53) | ||
| 25.00 | <10 | ||
| 12.5 | <10 | ||
| 7,8-Dihydroxy-flavone | Docking | 100.00 | 39.8 (0.31) |
| 50.00 | 34.54 (0.13) | ||
| 25.00 | <10 | ||
| 12.5 | <10 | ||
| Isokaempferide | Cluster | 100.00 | 49.98 (22.38) |
| 50.00 | 40.06 (27.12) | ||
| 25.00 | 19.69 (13.71) | ||
| 12.5 | <10 |
| Compound | Concentrations Tested (µg/mL) | Antiviral Activity against F-CoV | Antiviral Activity against B-CoV | Antiviral Activity against OC43 |
|---|---|---|---|---|
| Baicalein | 100 | TX | TX | TX |
| 50 | 66.73 | 99.60 | 99.99 | |
| 25 | <50 | 89.85 | 99.99 | |
| 12.5 | <50 | <50 | 99.98 | |
| 6.25 | <50 | <50 | <50 | |
| 3.125 | <50 | <50 | <50 | |
| 1.56 | <50 | <50 | <50 | |
| 0.78 | <50 | <50 | <50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, M.; Quaglio, D.; Calcaterra, A.; Ghirga, F.; Sorrentino, L.; Cammarone, S.; Fracella, M.; D’Auria, A.; Frasca, F.; Criscuolo, E.; et al. Natural Flavonoid Derivatives Have Pan-Coronavirus Antiviral Activity. Microorganisms 2023, 11, 314. https://doi.org/10.3390/microorganisms11020314
Mori M, Quaglio D, Calcaterra A, Ghirga F, Sorrentino L, Cammarone S, Fracella M, D’Auria A, Frasca F, Criscuolo E, et al. Natural Flavonoid Derivatives Have Pan-Coronavirus Antiviral Activity. Microorganisms. 2023; 11(2):314. https://doi.org/10.3390/microorganisms11020314
Chicago/Turabian StyleMori, Mattia, Deborah Quaglio, Andrea Calcaterra, Francesca Ghirga, Leonardo Sorrentino, Silvia Cammarone, Matteo Fracella, Alessandra D’Auria, Federica Frasca, Elena Criscuolo, and et al. 2023. "Natural Flavonoid Derivatives Have Pan-Coronavirus Antiviral Activity" Microorganisms 11, no. 2: 314. https://doi.org/10.3390/microorganisms11020314








