Dynamics of Hepatitis B Virus Covalently Closed Circular DNA: A Mini-Review
Abstract
:1. Introduction
2. The Term ‘cccDNA Half-Life’ Does Not Refer to an Intrinsic Property of cccDNA
3. Major Factors That Affect cccDNA Half-Life
3.1. Noncytopathic Effects Contribute to cccDNA Degradation in Acute Infection
3.2. Hepatocyte Turnover Is a Major Factor Promoting cccDNA Decay
3.2.1. Evidence That Hepatocyte Turnover Promotes cccDNA Decay
3.2.2. Effect of Cell Division on cccDNA
3.2.3. Hepatocyte Turnover Rate

4. HBV Genome Recycling and cccDNA Replenishment
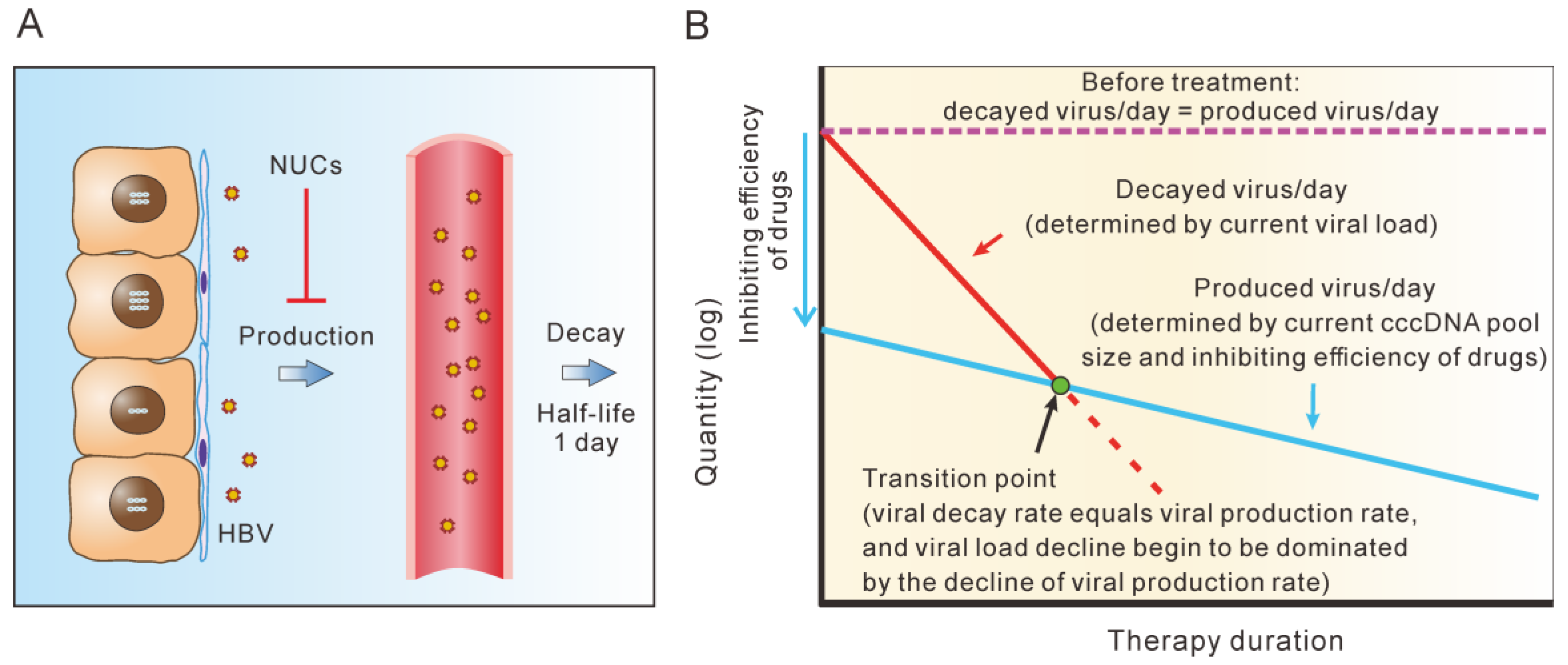
5. cccDNA Decay Dominates the Second Phase of Serum HBV Decline during Short-Term NUC Therapy
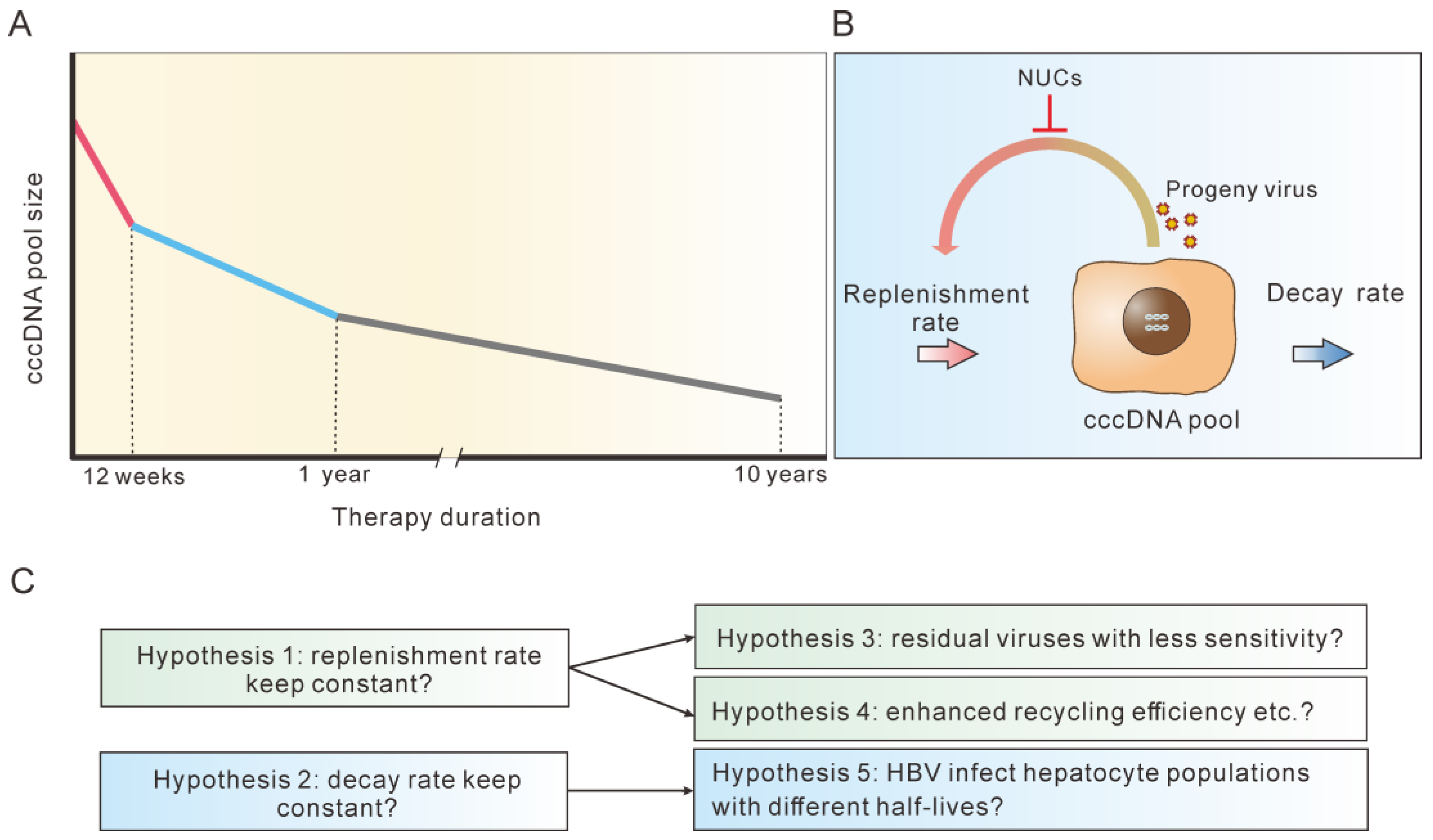
6. A Significant Variation in cccDNA Half-life Was Observed during Different Terms of NUC Therapy
7. Possible Explanations for the Multi-Phasic Decline of cccDNA during NUC Therapy
8. Considering CHB Treatments in Light of cccDNA Dynamics
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
List of Abbreviations
References
- Lok, A.S.; Zoulim, F.; Dusheiko, G.; Ghany, M.G. Hepatitis B cure: From discovery to regulatory approval. Hepatology 2017, 66, 1296–1313. [Google Scholar] [CrossRef] [Green Version]
- Ning, Q.; Wu, D.; Wang, G.-Q.; Ren, H.; Gao, Z.-L.; Hu, P.; Han, M.-F.; Wang, Y.; Zhang, W.-H.; Lu, F.-M.; et al. Roadmap to functional cure of chronic hepatitis B: An expert consensus. J. Viral Hepat. 2019, 26, 1146–1155. [Google Scholar] [CrossRef]
- Wong, G.L.H.; Gane, E.; Lok, A.S.F. How to achieve functional cure of HBV: Stopping NUCs, adding interferon or new drug de-velopment? J. Hepatol. 2022, 76, 1249–1262. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ploss, A. Mechanism of Hepatitis B Virus cccDNA Formation. Viruses 2021, 13, 1463. [Google Scholar] [CrossRef]
- Xia, Y.; Guo, H. Hepatitis B virus cccDNA: Formation, regulation and therapeutic potential. Antivir. Res. 2020, 180, 104824. [Google Scholar] [CrossRef]
- Hong, X.; Kim, E.S.; Guo, H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology 2017, 66, 2066–2077. [Google Scholar] [CrossRef] [Green Version]
- Block, T.M.; Alter, H.; Brown, N.; Brownstein, A.; Brosgart, C.; Chang, K.-M.; Chen, P.-J.; Cohen, C.; El-Serag, H.; Feld, J.; et al. Research priorities for the discovery of a cure for chronic hepatitis B: Report of a workshop. Antivir. Res. 2018, 150, 93–100. [Google Scholar] [CrossRef]
- Martinez, M.G.; Boyd, A.; Combe, E.; Testoni, B.; Zoulim, F. Covalently closed circular DNA: The ultimate therapeutic target for curing HBV infections. J. Hepatol. 2021, 75, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.G.; Baumert, T.F.; Boni, C.; Gane, E.; Levrero, M.; Lok, A.S.; Maini, M.K.; Terrault, N.A.; Zoulim, F. The scientific basis of combination therapy for chronic hepatitis B functional cure. Nat. Rev. Gastroenterol. Hepatol. 2023, 1–16. [Google Scholar] [CrossRef]
- Dusheiko, G.; Agarwal, K.; Maini, M.K. New Approaches to Chronic Hepatitis B. N. Engl. J. Med. 2023, 388, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Degasperi, E.; Anolli, M.P.; Lampertico, P. Towards a Functional Cure for Hepatitis B Virus: A 2022 Update on New Antiviral Strategies. Viruses 2022, 14, 2404. [Google Scholar] [CrossRef]
- Zoulim, F. Inhibition of hepatitis B virus gene expression: A step towards functional cure. J. Hepatol. 2018, 68, 386–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, L.-Y.; Cheung, K.-S.; Fung, J.; Seto, W.-K.; Yuen, M.-F. New strategies for the treatment of chronic hepatitis B. Trends Mol. Med. 2022, 28, 742–757. [Google Scholar] [CrossRef]
- Fanning, G.C.; Zoulim, F.; Hou, J.; Bertoletti, A. Therapeutic strategies for hepatitis B virus infection: Towards a cure. Nat. Rev. Drug Discov. 2019, 18, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Naggie, S.; Lok, A.S. New Therapeutics for Hepatitis B: The Road to Cure. Annu. Rev. Med. 2021, 72, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhang, E.; Gao, S.; Xiong, Y.; Lu, M. Toward a Functional Cure for Hepatitis B: The Rationale and Challenges for Therapeutic Targeting of the B Cell Immune Response. Front. Immunol. 2019, 10, 2308. [Google Scholar] [CrossRef]
- Nowak, M.A.; Bonhoeffer, S.; Hill, A.M.; Boehme, R.; Thomas, H.C.; McDade, H. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 1996, 93, 4398–4402. [Google Scholar] [CrossRef] [Green Version]
- Boettler, T.; Gill, U.S.; Allweiss, L.; Pollicino, T.; Tavis, J.E.; Zoulim, F. Assessing immunological and virological responses in the liver: Implications for the cure of chronic hepatitis B virus infection. JHEP Rep. 2022, 4, 100480. [Google Scholar] [CrossRef]
- Murray, J.M.; Wieland, S.F.; Purcell, R.H.; Chisari, F.V. Dynamics of hepatitis B virus clearance in chimpanzees. Proc. Natl. Acad. Sci. USA 2005, 102, 17780–17785. [Google Scholar] [CrossRef] [Green Version]
- Boyd, A.; Lacombe, K.; Lavocat, F.; Maylin, S.; Miailhes, P.; Lascoux-Combe, C.; Delaugerre, C.; Girard, P.M.; Zoulim, F. Decay of ccc-DNA marks per-sistence of intrahepatic viral DNA synthesis under tenofovir in HIV-HBV co-infected patients. J. Hepatol. 2016, 65, 683–691. [Google Scholar] [CrossRef]
- Guidotti, L.G.; Ishikawa, T.; Hobbs, M.V.; Matzke, B.; Schreiber, R.; Chisari, F.V. Intracellular Inactivation of the Hepatitis B Virus by Cytotoxic T Lymphocytes. Immunity 1996, 4, 25–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Stadler, D.; Lucifora, J.; Reisinger, F.; Webb, D.; Hösel, M.; Michler, T.; Wisskirchen, K.; Cheng, X.; Zhang, K.; et al. Interferon-gamma and Tumor Necrosis Factor-alpha Produced by T Cells Reduce the HBV Persistence Form, cccDNA, Without Cytolysis. Gastroenterology 2016, 150, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, L.G.; Rochford, R.; Chung, J.; Shapiro, M.; Purcell, R.; Chisari, F.V. Viral Clearance Without Destruction of Infected Cells During Acute HBV Infection. Science 1999, 284, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Mason, W.S.; Xu, C.; Low, H.C.; Saputelli, J.; Aldrich, C.E.; Scougall, C.; Grosse, A.; Colonno, R.; Litwin, S.; Jilbert, A.R. The amount of hepatocyte turnover that oc-curred during resolution of transient hepadnavirus infections was lower when virus replication was inhibited with entecavir. J. Virol. 2009, 83, 1778–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidotti, L.G.; Ando, K.; Hobbs, M.V.; Ishikawa, T.; Runkel, L.; Schreiber, R.D.; Chisari, F.V. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. USA 1994, 91, 3764–3768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Campagna, M.; Qi, Y.; Zhao, X.; Guo, F.; Xu, C.; Li, S.; Li, W.; Block, T.M.; Chang, J.; et al. Alpha-Interferon Suppresses Hepadnavirus Transcription by Altering Epigenetic Modification of cccDNA Minichromosomes. PLoS Pathog. 2013, 9, e1003613. [Google Scholar] [CrossRef] [Green Version]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T.; et al. Specific and Nonhepatotoxic Degradation of Nuclear Hepatitis B Virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef]
- Fourel, I.; Cullen, J.M.; Saputelli, J.; Aldrich, C.E.; Schaffer, P.; Averett, D.R.; Pugh, J.; Mason, W.S. Evidence that hepatocyte turnover is required for rapid clearance of duck hepatitis B virus during antiviral therapy of chronically infected ducks. J. Virol. 1994, 68, 8321–8330. [Google Scholar] [CrossRef] [Green Version]
- Addison, W.R.; Walters, K.A.; Wong, W.W.; Wilson, J.S.; Madej, D.; Jewell, L.D.; Tyrrell, D.L. Half-life of the duck hepatitis B virus co-valently closed circular DNA pool in vivo following inhibition of viral replication. J. Virol. 2002, 76, 6356–6363. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Yamamoto, T.; Cullen, J.; Saputelli, J.; Aldrich, C.E.; Miller, D.S.; Litwin, S.; Furman, P.A.; Jilbert, A.; Mason, W.S. Kinetics of Hepadnavirus Loss from the Liver during Inhibition of Viral DNA Synthesis. J. Virol. 2001, 75, 311–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohne, F.; Chmielewski, M.; Ebert, G.; Wiegmann, K.; Kürschner, T.; Schulze, A.; Urban, S.; Krönke, M.; Abken, H.; Protzer, U. T Cells Redirected Against Hepatitis B Virus Surface Proteins Eliminate Infected Hepatocytes. Gastroenterology 2008, 134, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Krebs, K.; Böttinger, N.; Huang, L.; Chmielewski, M.; Arzberger, S.; Gasteiger, G.; Jäger, C.; Schmitt, E.; Bohne, F.; Aichler, M.; et al. T Cells Expressing a Chimeric Antigen Receptor That Binds Hepatitis B Virus Envelope Proteins Control Virus Replication in Mice. Gastroenterology 2013, 145, 456–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisskirchen, K.; Kah, J.; Malo, A.; Asen, T.; Volz, T.; Allweiss, L.; Wettengel, J.M.; Lütgehetmann, M.; Urban, S.; Bauer, T.; et al. T cell receptor grafting allows virological control of hepatitis B virus infection. J. Clin. Investig. 2019, 129, 2932–2945. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Chakraborty, A.; Chou, W.-M.; Hasreiter, J.; Wettengel, J.M.; Stadler, D.; Bester, R.; Asen, T.; Zhang, K.; Wisskirchen, K.; et al. Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J. Hepatol. 2018, 69, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.L.; Chen, M.L.; Wu, Y.C.; Tsai, K.N.; Huang, C.C.; Hu, C.P.; Jeng, K.S.; Chou, Y.C.; Chang, C. Dynamics of HBV cccDNA expression and tran-scription in different cell growth phase. J. Biomed. Sci. 2011, 18, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, T.; Zehnder, B.; Wettengel, J.M.; Zhang, H.; Coulter, S.; Ho, V.; Douglas, M.W.; Protzer, U.; George, J.; Urban, S. Mitosis of hepatitis B virus-infected cells in vitro results in uninfected daughter cells. JHEP Rep. 2022, 4, 100514. [Google Scholar] [CrossRef]
- Li, M.; Sohn, J.A.; Seeger, C. Distribution of Hepatitis B Virus Nuclear DNA. J. Virol. 2018, 92, e01391-17. [Google Scholar] [CrossRef] [Green Version]
- Dandri, M.; Burda, M.R.; Will, H.; Petersen, J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus repli-cation by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology 2000, 32, 139–146. [Google Scholar] [CrossRef]
- Lutgehetmann, M.; Volz, T.; Köpke, A.; Broja, T.; Tigges, E.; Lohse, A.W.; Fuchs, E.; Murray, J.M.; Petersen, J.; Dandri, M. In vivo proliferation of hepadnavirus-infected hepatocytes induces loss of covalently closed circular DNA in mice. Hepatology 2010, 52, 16–24. [Google Scholar] [CrossRef]
- Allweiss, L.; Volz, T.; Giersch, K.; Kah, J.; Raffa, G.; Petersen, J.; Lohse, A.W.; Beninati, C.; Pollicino, T.; Urban, S.; et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut 2018, 67, 542–552. [Google Scholar] [CrossRef]
- Macdonald, R.A. “Lifespan” of liver cells. Autoradio-graphic study using tritiated thymidine in normal, cirrhotic, and partially hepatectomized rats. Arch. Intern. Med. 1961, 107, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Magami, Y.; Azuma, T.; Inokuchi, H.; Kokuno, S.; Moriyasu, F.; Kawai, K.; Hattori, T. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver Int. 2002, 22, 419–425. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Pu, W.; Liu, X.; Zhang, Z.; Han, M.; Li, Y.; Huang, X.; Han, X.; Li, Y.; Liu, K.; et al. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science 2021, 371, eabc4346. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wang, Y.G.; Jia, Y.; Li, L.; Yoon, J.; Zhang, S.; Wang, Z.; Zhang, Y.; Zhu, M.; Sharma, T.; et al. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science 2021, 371, eabb1625. [Google Scholar] [CrossRef] [PubMed]
- Celis, J.E.; Celis, A. Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: Subdivision of S phase. Proc. Natl. Acad. Sci. USA 1985, 82, 3262–3266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, R.; Macdonald-Bravo, H. Changes in the nuclear distribution of cyclin (PCNA) but not its synthesis depend on DNA replication. EMBO J. 1985, 4, 655–661. [Google Scholar] [CrossRef]
- Bravo, R.; Macdonald-Bravo, H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: Association with DNA replication sites. J. Cell Biol. 1987, 105, 1549–1554. [Google Scholar] [CrossRef] [Green Version]
- Mancini, R.; Marucci, L.; Benedetti, A.; Jezequel, A.-M.; Orlandi, F. Immunohistochemical analysis of S-phase cells in normal human and rat liver by PC10 monoclonal antibody. Liver Int. 1994, 14, 57–64. [Google Scholar] [CrossRef]
- Nakajima, T.; Kagawa, K.; Ueda, K.; Ohkawara, T.; Kimura, H.; Kakusui, M.; Deguchi, T.; Okanoue, T.; Kashima, K.; Ashihara, T. Evaluation of hepatic proliferative activity in chronic liver diseases and hepatocellular carcinomas by proliferating cell nuclear antigen (PCNA) immunohisto-chemical staining of methanol-fixed tissues. J. Gastroenterol. 1994, 29, 450–454. [Google Scholar] [CrossRef]
- Mason, W.S.; Jilbert, A.R.; Litwin, S. Hepatitis B Virus DNA Integration and Clonal Expansion of Hepatocytes in the Chronically Infected Liver. Viruses 2021, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- Köck, J.; Rösler, C.; Zhang, J.-J.; Blum, H.E.; Nassal, M.; Thoma, C. Generation of Covalently Closed Circular DNA of Hepatitis B Viruses via Intracellular Recycling Is Regulated in a Virus Specific Manner. PLoS Pathog. 2010, 6, e1001082. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Jiang, D.; Zhou, T.; Cuconati, A.; Block, T.M.; Guo, J.-T. Characterization of the Intracellular Deproteinized Relaxed Circular DNA of Hepatitis B Virus: An Intermediate of Covalently Closed Circular DNA Formation. J. Virol. 2007, 81, 12472–12484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Hu, J. Formation of Hepatitis B Virus Covalently Closed Circular DNA: Removal of Genome-Linked Protein. J. Virol. 2007, 81, 6164–6174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostyushev, D.; Kostyusheva, A.; Brezgin, S.; Ponomareva, N.; Zakirova, N.F.; Egorshina, A.; Yanvarev, D.V.; Bayurova, E.; Sudina, A.; Goptar, I.; et al. Depleting hepatitis B virus relaxed circular DNA is necessary for resolution of infection by CRISPR-Cas9. Mol. Ther. Nucleic Acids 2023, 31, 482–493. [Google Scholar] [CrossRef]
- Tu, T.; Zehnder, B.; Qu, B.; Urban, S. De novo synthesis of hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep. 2021, 3, 100195. [Google Scholar] [CrossRef]
- Volz, T.; Allweiss, L.; Ben Ḿbarek, M.; Warlich, M.; Lohse, A.W.; Pollok, J.M.; Alexandrov, A.; Urban, S.; Petersen, J.; Lütgehetmann, M.; et al. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J. Hepatol. 2013, 58, 861–867. [Google Scholar] [CrossRef]
- Tsiang, M.; Rooney, J.F.; Toole, J.J.; Gibbs, C.S. Biphasic clearance kinetics of hepatitis B virus from patients during adefovir dipivoxil therapy. Hepatology 1999, 29, 1863–1869. [Google Scholar] [CrossRef]
- Wolters, L.M.; E Hansen, B.; Niesters, H.G.; DeHertogh, D.; de Man, R.A. Viral dynamics during and after entecavir therapy in patients with chronic hepatitis B. J. Hepatol. 2002, 37, 137–144. [Google Scholar] [CrossRef]
- Goncalves, A.; Lemenuel-Diot, A.; Cosson, V.; Jin, Y.; Feng, S.; Bo, Q.; Guedj, J. What drives the dynamics of HBV RNA during treatment? J. Viral. Hepat. 2021, 28, 383–392. [Google Scholar] [CrossRef]
- Wolters, L.; Hansen, B.; Niesters, H.; Zeuzem, S.; Schalm, S.; De, M.R.; De Man, R.A. Viral dynamics in chronic hepatitis B patients during lamivudine therapy. Liver Int. 2002, 22, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Werle-Lapostolle, B.; Bowden, S.; Locarnini, S.; Wursthorn, K.; Petersen, J.; Lau, G.; Trepo, C.; Marcellin, P.; Goodman, Z.; Delaney, W.E., IV; et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 2004, 126, 1750–1758. [Google Scholar] [CrossRef] [Green Version]
- Wursthorn, K.; Lutgehetmann, M.; Dandri, M.; Volz, T.; Buggisch, P.; Zollner, B.; Longerich, T.; Schirmacher, P.; Metzler, F.; Zankel, M.; et al. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 2006, 44, 675–684. [Google Scholar] [CrossRef]
- Takkenberg, B.; Terpstra, V.; Zaaijer, H.; Weegink, C.; Dijkgraaf, M.; Jansen, P.; Beld, M.; Reesink, H. Intrahepatic response markers in chronic hepatitis B patients treated with peginterferon alpha-2a and adefovir. J. Gastroenterol. Hepatol. 2011, 26, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Bowden, S.; Locarnini, S.; Chang, T.-T.; Chao, Y.-C.; Han, K.-H.; Gish, R.G.; de Man, R.A.; Yu, M.; Llamoso, C.; Tang, H. Covalently closed-circular hepatitis B virus DNA reduction with entecavir or lamivudine. World J. Gastroenterol. 2015, 21, 4644–4651. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.K.-H.; Yuen, M.-F.; Ngai, V.W.-S.; Fung, J.; Lai, C.-L. One-Year Entecavir or Lamivudine Therapy Results in Reduction of Hepatitis B Virus Intrahepatic Covalently Closed Circular DNA Levels. Antivir. Ther. 2006, 11, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.K.-H.; Seto, W.-K.; Fung, J.; Ip, P.; Huang, F.-Y.; Lai, C.-L.; Yuen, M.-F. Reduction of Hepatitis B Surface Antigen and Covalently Closed Circular DNA by Nucleos(t)ide Analogues of Different Potency. Clin. Gastroenterol. Hepatol. 2013, 11, 1004–1010.e1. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhu, Y.Y.; Chen, J.; Liu, Y.R.; You, J.; Dong, J.; Zeng, D.W.; Gao, L.Y.; Chen, L.H.; Jiang, J.J. Decline in intrahepatic cccDNA and increase in immune cell reactivity after 12 weeks of antiviral treatment were associated with HBeAg loss. J. Viral Hepat. 2014, 21, 909–916. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Liao, H.; Deng, Z.; Bian, D.; Ren, Y.; Yu, G.; Jiang, Y.; Bai, L.; Liu, S.; et al. Serum HBV DNA plus RNA reflecting cccDNA level before and during NAs treatment in HBeAg positive CHB patients. Int. J. Med. Sci. 2022, 19, 858–866. [Google Scholar] [CrossRef]
- Lai, C.-L.; Wong, D.; Ip, P.; Kopaniszen, M.; Seto, W.-K.; Fung, J.; Huang, F.-Y.; Lee, B.; Cullaro, G.; Chong, C.K.; et al. Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B. J. Hepatol. 2017, 66, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Zhou, B.; Cai, D.; Zong, Y.; Wu, Y.; Liu, S.; Mercier, A.; Guo, H.; Hou, J.; Colonno, R.; et al. Rapid Turnover of Hepatitis B Virus Covalently Closed Circular DNA Indicated by Monitoring Emergence and Reversion of Signature-Mutation in Treated Chronic Hepatitis B Patients. Hepatology 2021, 73, 41–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [Green Version]
- Payen, V.L.; Lavergne, A.; Sarika, N.A.; Colonval, M.; Karim, L.; Deckers, M.; Najimi, M.; Coppieters, W.; Charloteaux, B.; Sokal, E.M.; et al. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021, 3, 100278. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, A.; Grudda, T.; Ribeiro, R.M.; Saad, Y.S.; Hwang, H.S.; Quinn, J.; Murphy, M.; Ward, K.; Sterling, R.K.; Zhang, Y.; et al. Single hepatocytes show persistence and transcriptional inactivity of hepatitis B. J. Clin. Investig. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Balagopal, A.; Hwang, H.S.; Grudda, T.; Quinn, J.; Sterling, R.K.; Sulkowski, M.S.; Thio, C.L. Single Hepatocyte Hepatitis B Virus Transcriptional Landscape in HIV Coinfection. J. Infect. Dis. 2020, 221, 1462–1469. [Google Scholar] [CrossRef]
- Wieland, S.F.; Spangenberg, H.C.; Thimme, R.; Purcell, R.H.; Chisari, F.V. Expansion and contraction of the hepatitis B virus tran-scriptional template in infected chimpanzees. Proc. Natl. Acad. Sci. USA 2004, 101, 2129–2134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thimme, R.; Wieland, S.; Steiger, C.; Ghrayeb, J.; Reimann, K.A.; Purcell, R.H.; Chisari, F.V. CD8+ T Cells Mediate Viral Clearance and Disease Pathogenesis during Acute Hepatitis B Virus Infection. J. Virol. 2003, 77, 68–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nijampatnam, B.; Liotta, D.C. Recent advances in the development of HBV capsid assembly modulators. Curr. Opin. Chem. Biol. 2019, 50, 73–79. [Google Scholar] [CrossRef]
- Zoulim, F.; Zlotnick, A.; Buchholz, S.; Donaldson, E.; Fry, J.; Gaggar, A.; Hu, J.; Kann, M.; Lenz, O.; Lin, K.; et al. Nomenclature of HBV core protein-targeting antivirals. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 748–750. [Google Scholar] [CrossRef]
- Taverniti, V.; Ligat, G.; Debing, Y.; Kum, D.B.; Baumert, T.F.; Verrier, E.R. Capsid Assembly Modulators as Antiviral Agents against HBV: Molecular Mechanisms and Clinical Perspectives. J. Clin. Med. 2022, 11, 1349. [Google Scholar] [CrossRef]
- Viswanathan, U.; Mani, N.; Hu, Z.; Ban, H.; Du, Y.; Hu, J.; Chang, J.; Guo, J.-T. Targeting the multifunctional HBV core protein as a potential cure for chronic hepatitis B. Antivir. Res. 2020, 182, 104917. [Google Scholar] [CrossRef] [PubMed]
- Roca Suarez, A.A.; Testoni, B.; Zoulim, F. HBV 2021: New therapeutic strategies against an old foe. Liver Int. 2021, 41 (Suppl. 1), 15–23. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Urban, S. Virus entry and its inhibition to prevent and treat hepatitis B and hepatitis D virus infections. Curr. Opin. Virol. 2018, 30, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.; Choi, H.S.J.; Gehring, A.; Janssen, H.L.A. Getting to HBV cure: The promising paths forward. Hepatology 2022, 76, 233–250. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.W.-H.; Mak, L.-Y.; Seto, W.-K.; Yuen, M.-F. RNA interference as a novel treatment strategy for chronic hepatitis B infection. Clin. Mol. Hepatol. 2022, 28, 408–424. [Google Scholar] [CrossRef]
- Allweiss, L.; Giersch, K.; Pirosu, A.; Volz, T.; Muench, R.C.; Beran, R.K.; Urban, S.; Javanbakht, H.; Fletcher, S.P.; Lütgehetmann, M.; et al. Therapeutic shutdown of HBV transcripts promotes reappearance of the SMC5/6 complex and silencing of the viral genome in vivo. Gut 2022, 71, 372–381. [Google Scholar] [CrossRef]
- Van den Berg, F.; Limani, S.W.; Mnyandu, N.; Maepa, M.B.; Ely, A.; Arbuthnot, P. Advances with RNAi-Based Therapy for Hepatitis B Virus Infection. Viruses 2020, 12, 851. [Google Scholar] [CrossRef]
- Yuen, M.; Schiefke, I.; Yoon, J.; Ahn, S.H.; Heo, J.; Kim, J.H.; Chan, H.L.Y.; Yoon, K.T.; Klinker, H.; Manns, M.; et al. RNA Interference Therapy With ARC-520 Results in Prolonged Hepatitis B Surface Antigen Response in Patients with Chronic Hepatitis B Infection. Hepatology 2020, 72, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Yuen, M.-F.; Wong, D.K.-H.; Schluep, T.; Lai, C.-L.; Ferrari, C.; Locarnini, S.; Lo, R.C.-L.; Gish, R.G.; Hamilton, J.; Wooddell, C.I.; et al. Long-term serological, virological and histological responses to RNA inhibition by ARC-520 in Chinese chronic hepatitis B patients on entecavir treatment. Gut 2022, 71, 789–797. [Google Scholar] [CrossRef]
- Yuen, M.-F.; Locarnini, S.; Lim, T.H.; Strasser, S.I.; Sievert, W.; Cheng, W.; Thompson, A.J.; Given, B.D.; Schluep, T.; Hamilton, J.; et al. Combination treatments including the small-interfering RNA JNJ-3989 induce rapid and sometimes prolonged viral responses in patients with CHB. J. Hepatol. 2022, 77, 1287–1298. [Google Scholar] [CrossRef]
- Michler, T.; Kosinska, A.D.; Festag, J.; Bunse, T.; Su, J.; Ringelhan, M.; Imhof, H.; Grimm, D.; Steiger, K.; Mogler, C.; et al. Knockdown of Virus Antigen Expression Increases Therapeutic Vaccine Efficacy in High-Titer Hepatitis B Virus Carrier Mice. Gastroenterology 2020, 158, 1762–1775.e9. [Google Scholar] [CrossRef] [PubMed]
- Yuen, M.-F.; Heo, J.; Jang, J.-W.; Yoon, J.-H.; Kweon, Y.-O.; Park, S.-J.; Tami, Y.; You, S.; Yates, P.; Tao, Y.; et al. Safety, tolerability and antiviral activity of the antisense oligonucleotide bepirovirsen in patients with chronic hepatitis B: A phase 2 randomized controlled trial. Nat. Med. 2021, 27, 1725–1734. [Google Scholar] [CrossRef]
- Yuen, M.-F.; Lim, S.-G.; Plesniak, R.; Tsuji, K.; Janssen, H.L.; Pojoga, C.; Gadano, A.; Popescu, C.P.; Stepanova, T.; Asselah, T.; et al. Efficacy and Safety of Bepirovirsen in Chronic Hepatitis B Infection. N. Engl. J. Med. 2022, 387, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, M.; Pântea, V.; Placinta, G.; Moscalu, I.; Cebotarescu, V.; Cojuhari, L.; Jimbei, P.; Iarovoi, L.; Smesnoi, V.; Musteata, T.; et al. Safety and Efficacy of 48 Weeks REP 2139 or REP 2165, Tenofovir Disoproxil, and Pegylated Interferon Alfa-2a in Patients with Chronic HBV Infection Naive to Nu-cleos(t)ide Therapy. Gastroenterology 2020, 158, 2180–2194. [Google Scholar] [CrossRef]
- Belloni, L.; Allweiss, L.; Guerrieri, F.; Pediconi, N.; Volz, T.; Pollicino, T.; Petersen, J.; Raimondo, G.; Dandri, M.; Levrero, M. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromo-some. J. Clin. Investig. 2012, 122, 529–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, M.G.; Combe, E.; Inchauspe, A.; Mangeot, P.E.; Delberghe, E.; Chapus, F.; Neveu, G.; Alam, A.; Carter, K.; Testoni, B.; et al. CRISPR-Cas9 Targeting of Hepatitis B Virus Covalently Closed Circular DNA Generates Transcriptionally Active Episomal Variants. Mbio 2022, 13, e02888-21. [Google Scholar] [CrossRef]
- Lin, H.; Li, G.; Peng, X.; Deng, A.; Ye, L.; Shi, L.; Wang, T.; He, J. The Use of CRISPR/Cas9 as a Tool to Study Human Infectious Viruses. Front. Cell. Infect. Microbiol. 2021, 11, 590989. [Google Scholar] [CrossRef]
- Yang, Y.-C.; Yang, H.-C. Recent Progress and Future Prospective in HBV Cure by CRISPR/Cas. Viruses 2021, 14, 4. [Google Scholar] [CrossRef]
- Amin, O.E.; Colbeck, E.J.; Daffis, S.; Khan, S.; Ramakrishnan, D.; Pattabiraman, D.; Chu, R.; Micolochick Steuer, H.; Lehar, S.; Peiser, L.; et al. Therapeutic Potential of TLR8 Agonist GS-9688 (Selgantolimod) in Chronic Hepatitis B: Remodeling of Antiviral and Regulatory Mediators. Hepatology 2021, 74, 55–71. [Google Scholar] [CrossRef]
- Kayesh, M.E.H.; Kohara, M.; Tsukiyama-Kohara, K. Toll-Like Receptor Response to Hepatitis B Virus Infection and Potential of TLR Agonists as Immunomodulators for Treating Chronic Hepatitis B: An Overview. Int. J. Mol. Sci. 2021, 22, 10462. [Google Scholar] [CrossRef]
- Balsitis, S.; Gali, V.; Mason, P.J.; Chaniewski, S.; Levine, S.M.; Wichroski, M.J.; Feulner, M.; Song, Y.; Granaldi, K.; Loy, J.K.; et al. Safety and efficacy of anti-PD-L1 therapy in the woodchuck model of HBV infection. PLoS ONE 2018, 13, e0190058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrando-Martinez, S.; Huang, K.; Bennett, A.S.; Sterba, P.; Yu, L.; Suzich, J.A.; Janssen, H.L.; Robbins, S.H. HBeAg seroconversion is associated with a more effective PD-L1 blockade during chronic hepatitis B infection. JHEP Rep. 2019, 1, 170–178. [Google Scholar] [CrossRef] [Green Version]
- Feray, C.; Lopez-Labrador, F.X. Is PD-1 blockade a potential therapy for HBV? JHEP Rep. 2019, 1, 142–144. [Google Scholar] [CrossRef] [Green Version]
- Whitacre, D.C.; Peters, C.J.; Sureau, C.; Nio, K.; Li, F.; Su, L.; Jones, J.E.; Isogawa, M.; Sallberg, M.; Frelin, L.; et al. Designing a therapeutic hepatitis B vaccine to circumvent immune tolerance. Hum. Vaccines Immunother. 2020, 16, 251–268. [Google Scholar] [CrossRef]
- Lim, S.G.; Agcaoili, J.; De Souza, N.N.A.; Chan, E. Therapeutic vaccination for chronic hepatitis B: A systematic review and me-ta-analysis. J. Viral. Hepat. 2019, 26, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Cargill, T.; Barnes, E. Therapeutic vaccination for treatment of chronic hepatitis B. Clin. Exp. Immunol. 2021, 205, 106–118. [Google Scholar] [CrossRef]
- Stasi, C.; Silvestri, C.; Voller, F. Hepatitis B vaccination and immunotherapies: An update. Clin. Exp. Vaccine Res. 2020, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kosinska, A.D.; Moeed, A.; Kallin, N.; Festag, J.; Su, J.; Steiger, K.; Michel, M.L.; Protzer, U.; Knolle, P.A. Synergy of therapeutic heterologous prime-boost hepatitis B vaccination with CpG-application to improve immune control of persistent HBV infection. Sci. Rep. 2019, 9, 10808. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.-Y.; Guo, X.-R.; Wu, Y.; Kang, X.-Z.; Zheng, Q.-B.; Qi, R.-Y.; Chen, B.-B.; Lan, Y.; Wei, M.; Wang, S.-J.; et al. A unique B cell epitope-based particulate vaccine shows effective suppression of hepatitis B surface antigen in mice. Gut 2020, 69, 343–354. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.R.; Yang, H.C.; Kuo, Y.T.; Liu, C.J.; Yang, T.Y.; Sung, K.C.; Lin, Y.Y.; Wang, H.Y.; Wang, C.C.; Shen, Y.C.; et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol. Ther. Nucleic Acids 2014, 3, e186. [Google Scholar] [CrossRef]
- Seeger, C.; Sohn, J.A. Targeting Hepatitis B Virus With CRISPR/Cas. Mol. Ther. Nucleic Acids 2014, 3, e216. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-C.; Chen, Y.-H.; Kao, J.-H.; Ching, C.; Liu, I.-J.; Wang, C.-C.; Tsai, C.-H.; Wu, F.-Y.; Liu, C.-J.; Chen, P.-J.; et al. Permanent Inactivation of HBV Genomes by CRISPR/Cas9-Mediated Non-cleavage Base Editing. Mol. Ther. Nucleic Acids 2020, 20, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.Y.; Sun, S.; Liang, Y.; Xu, H.; Zhang, C.; Zhang, M.; Wang, F.S.; Fu, Y.-X.; Peng, H. Engineered anti-PDL1 with IFNalpha targets both im-munoinhibitory and activating signals in the liver to break HBV immune tolerance. Gut 2022. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, Q.; Zhang, J.D.; Zhang, Y.; Ni, X.; Xiang, K.; Jiang, J.; Li, B.; Yu, Y.; Hu, H.; et al. Discovery of a first-in-class orally available HBV cccDNA inhibitor. J. Hepatol. 2022. [Google Scholar] [CrossRef] [PubMed]


| Therapy | Sample Size | Observation Period | Half-Life of Serum Virion | Half-Life of Infected Cells * | Reference |
|---|---|---|---|---|---|
| LAM | 45 | 28 days | 1 day | 16 days (10–100 days) | [17] |
| ADV | 13 | 12 weeks | 1.1 days | 18 days (11–30 days) | [58] |
| LAM | 21 | 4 weeks | 17 h | 7 days | [61] |
| ETV | 10 | 4 weeks | 16 h | 10.7 days | [59] |
| Therapy | Sample Size (Patients) | Observation Period | cccDNA Reduction | cccDNA Half-Life | Reference |
|---|---|---|---|---|---|
| ADV | 22 | 48 weeks | 0.8 log | [62] | |
| PEG-IFNα-2b + ADV | 26 | 48 weeks | 2.4 log | [63] | |
| PEG-IFNα-2a + ADV | 40 | 48 weeks | 1.03 log (HBeAg (+)) 0.44 log (HBeAg (−)) | [64] | |
| ETV or LAM | 305 | 48 weeks | 0.9 log (ETV) 0.7 log (LAM) | [65] | |
| ETV | 40 | 48 weeks | 1 log | [66] | |
| NUC | 117 | 52 weeks | 0.93 log | [67] | |
| ADV | 15 | 12 weeks | 0.65 log | [68] | |
| ETV or ADV | 54 | 60 months | 1.56 log | [69] | |
| TDF | 27 | 6.9 years | 8.6 months (HBeAg+) 26.2 months (HBeAg−) | [20] | |
| NUC | 43 | 72–145 months | 1.03 log in the 1st year, 2.94 log in 2–10 year | [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.-L.; Huang, A.-L. Dynamics of Hepatitis B Virus Covalently Closed Circular DNA: A Mini-Review. Microorganisms 2023, 11, 600. https://doi.org/10.3390/microorganisms11030600
Hu J-L, Huang A-L. Dynamics of Hepatitis B Virus Covalently Closed Circular DNA: A Mini-Review. Microorganisms. 2023; 11(3):600. https://doi.org/10.3390/microorganisms11030600
Chicago/Turabian StyleHu, Jie-Li, and Ai-Long Huang. 2023. "Dynamics of Hepatitis B Virus Covalently Closed Circular DNA: A Mini-Review" Microorganisms 11, no. 3: 600. https://doi.org/10.3390/microorganisms11030600






