Sessile Lifestyle Offers Protection against Copper Stress in Saccharolobus solfataricus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Planktonic Growth Conditions
2.2. Microtitration Plate Assays
2.3. Epifluorescence Microscopy and Confocal Laser Microscopy
2.4. Adhesion Assays
2.5. Total RNA Extraction and cDNA Synthesis
2.6. Primer Design and Real-Time RT-PCR
3. Results
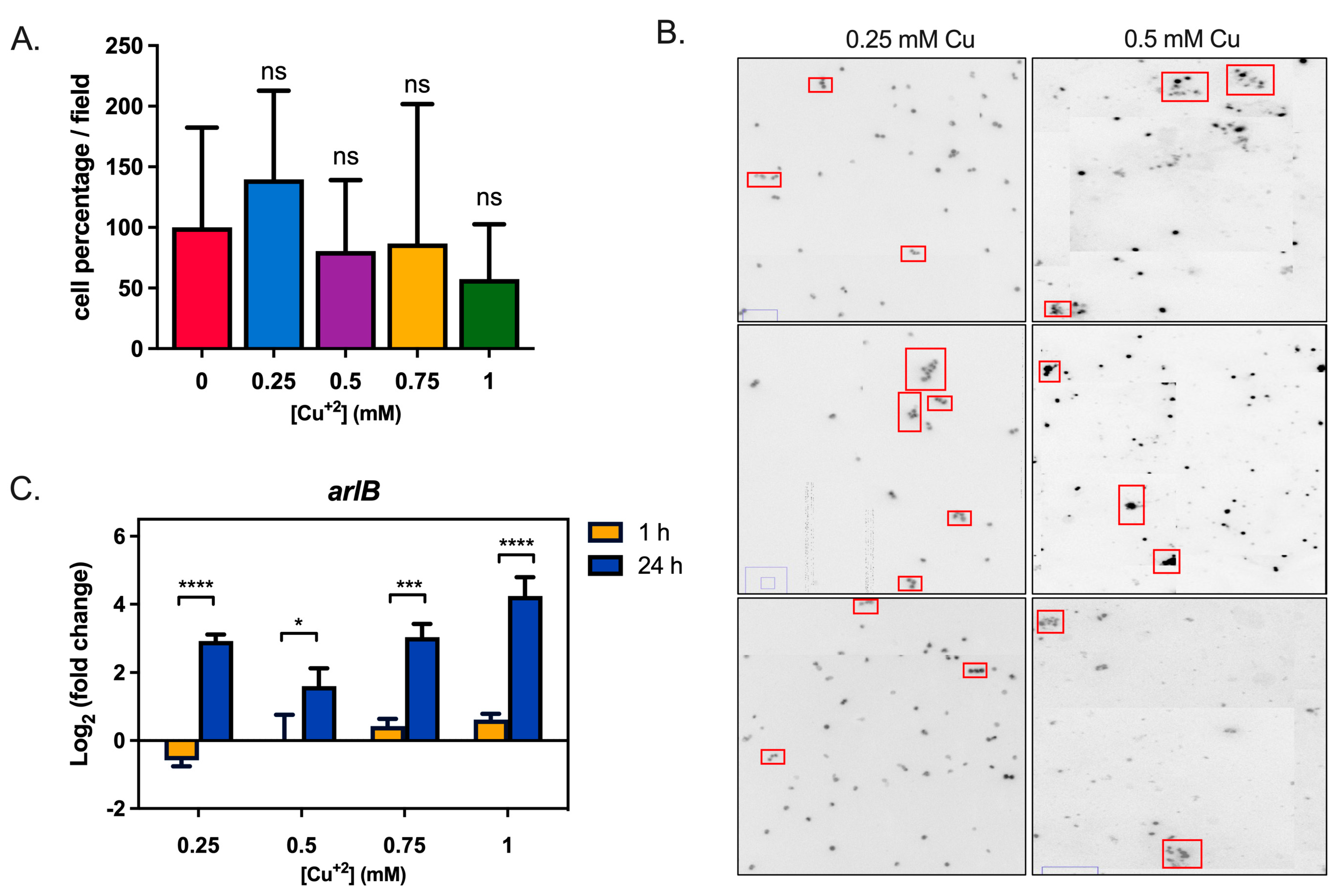
3.1. Copper Induces Biofilm Formation in S. solfataricus M16
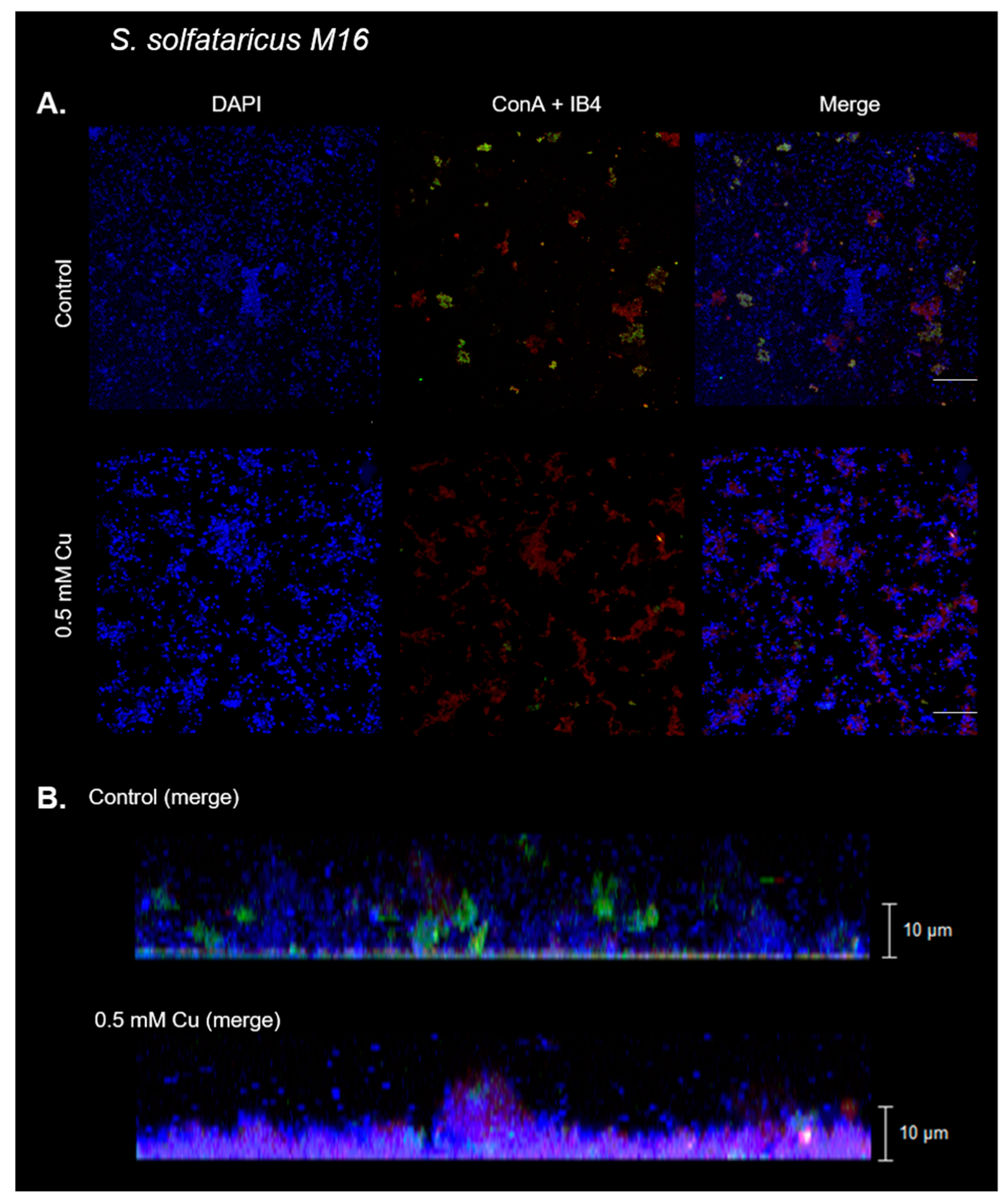
3.2. Cu Affects Biofilm Morphology
3.3. Cu Influences Archaellum Expression and Cell Adherence
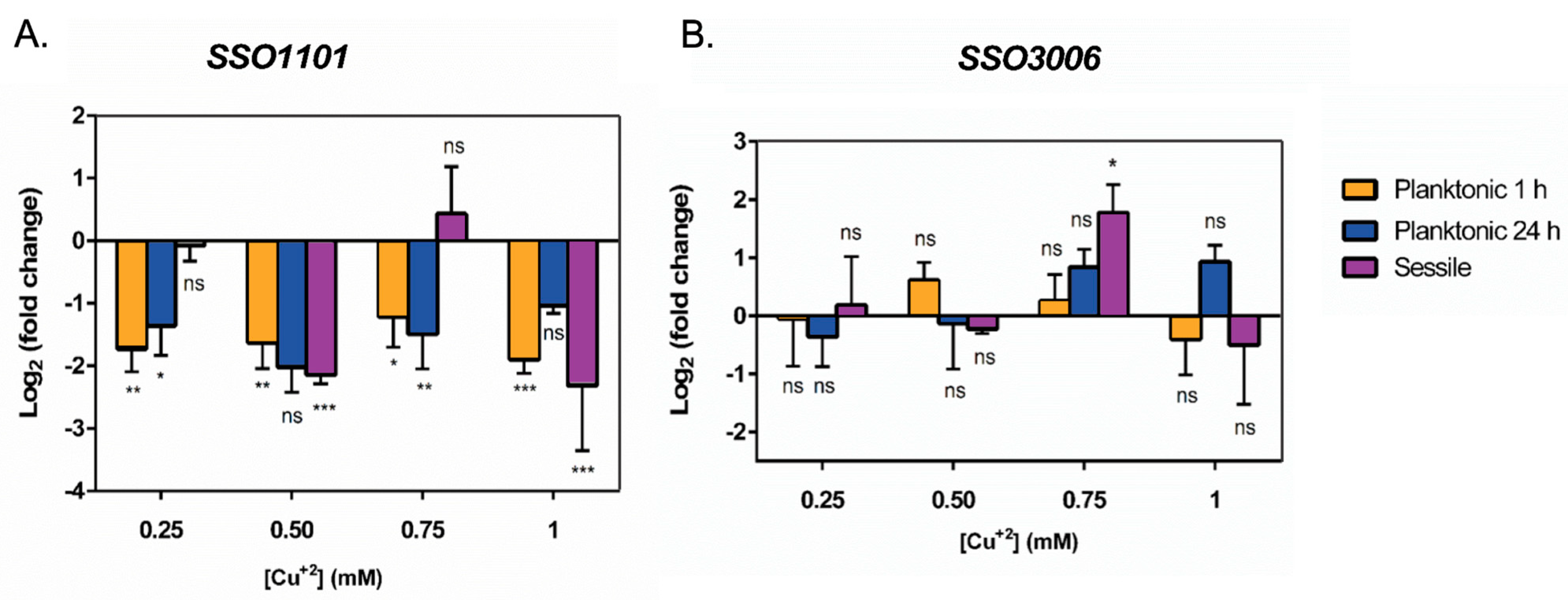
3.4. Transcriptional Levels of Genes Related to Biofilm in the Presence of Cu
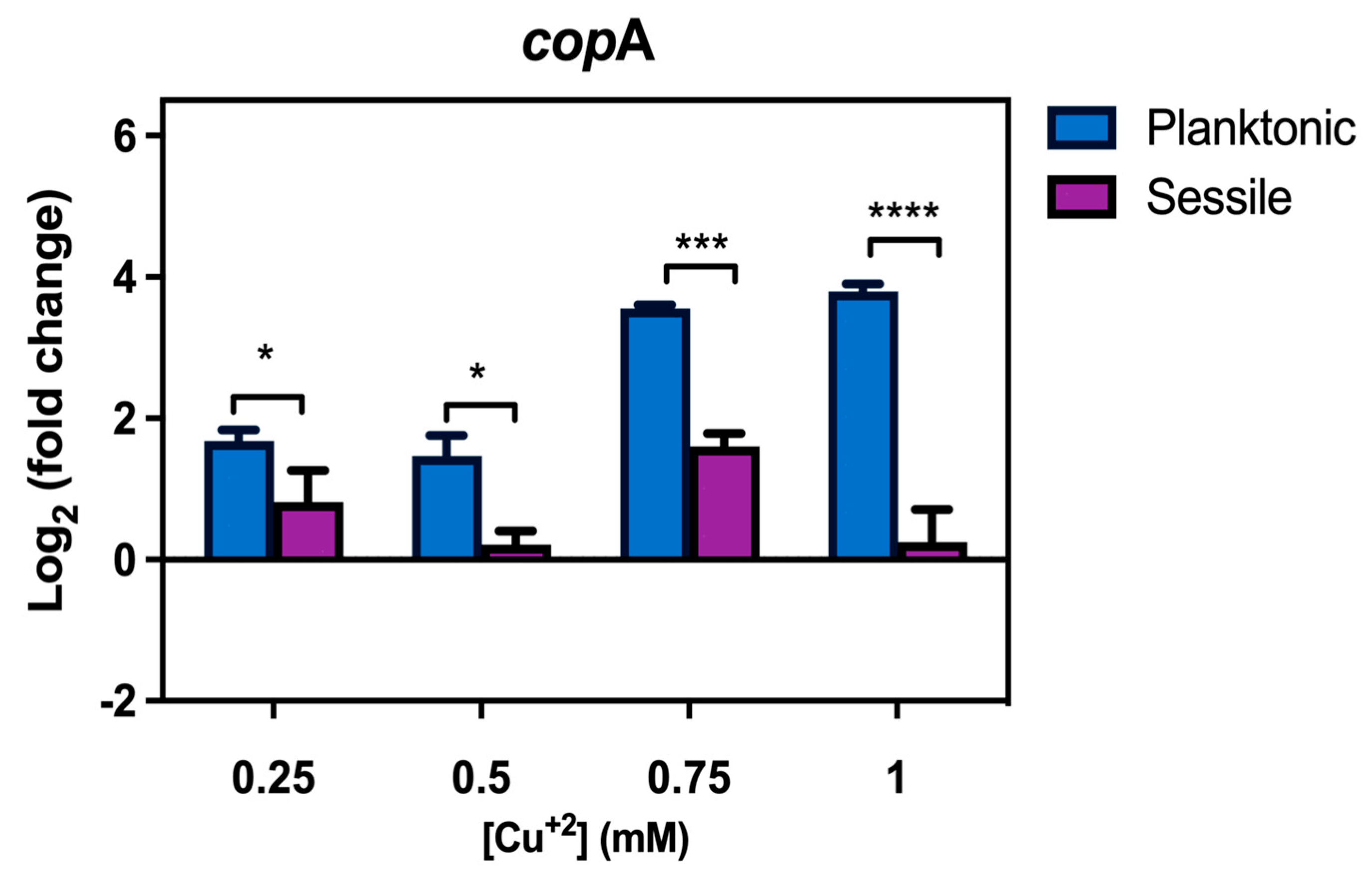
3.5. Transcriptional Levels of CopA Are Higher in Planktonic Than in Biofilm Lifestyle
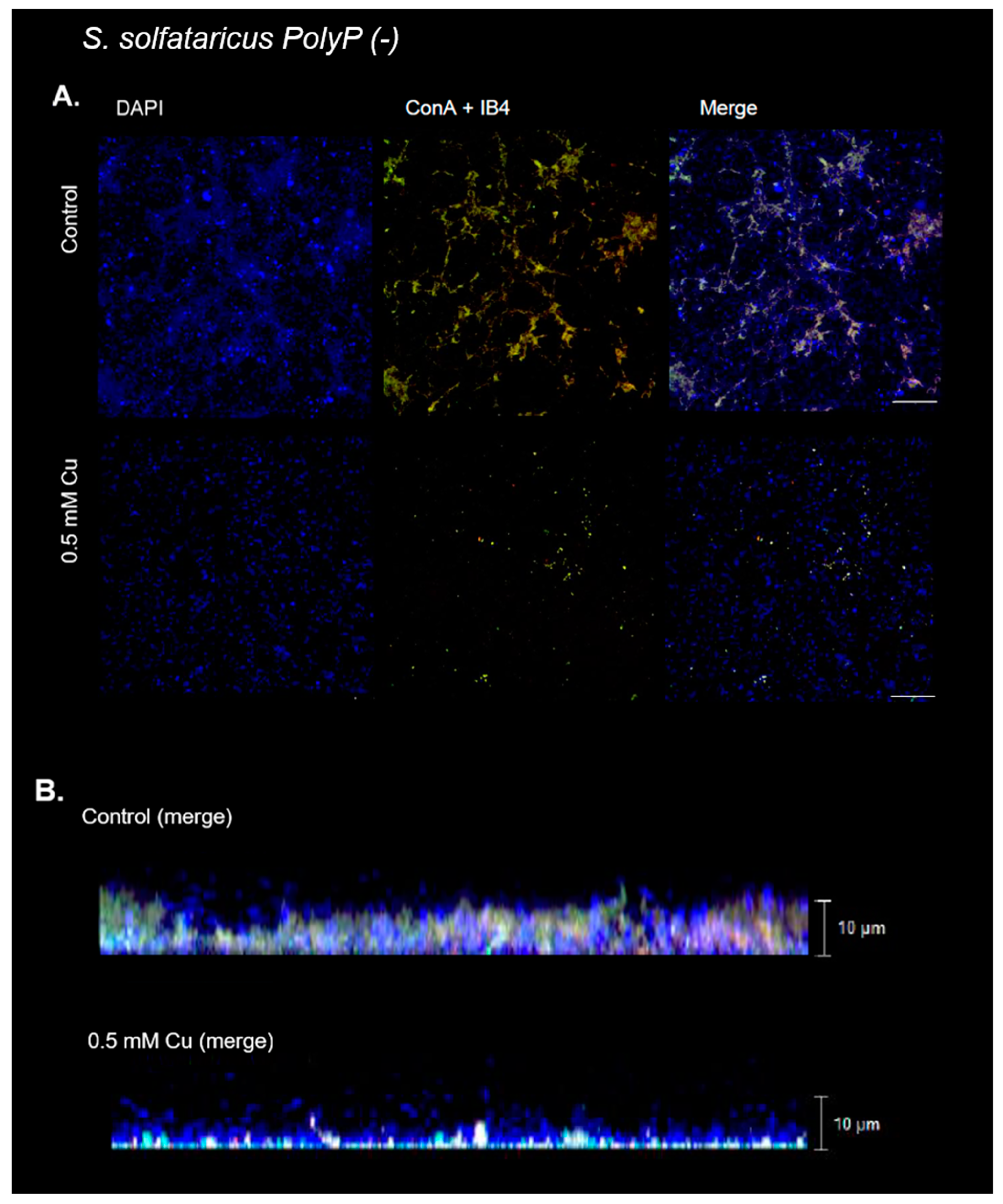
3.6. Cu Did Not Induce Biofilm Formation in a PolyP (-) Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Orell, A.; Guiliani, N. Biofilm Formation by Acidophile Bacteria and Archaea. In Acidophiles: Life in Extremely Acidic Environments; Johnson, D.B., Quatrini, R., Eds.; Caister Academic Press: Poole, UK, 2016; pp. 139–152. [Google Scholar] [CrossRef]
- Fröls, S. Archaeal biofilms: Widespread and complex. Biochem. Soc. Trans. 2013, 41, 393–398. [Google Scholar] [CrossRef]
- Orell, A.; Fröls, S.; Albers, S.V. Archaeal biofilms: The great unexplored. Annu. Rev. Microbiol. 2013, 67, 337–354. [Google Scholar] [CrossRef]
- Rinker, K.D.; Kelly, R.M. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: Development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence of biofilm formation. Appl. Environ. Microbiol. 1996, 62, 4478–4485. [Google Scholar] [CrossRef]
- Koerdt, A.; Gödeke, J.; Berger, J.; Thormann, K.M.; Albers, S.V. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 2010, 5, e14104. [Google Scholar] [CrossRef]
- Koerdt, A.; Orell, A.; Pham, T.K.; Mukherjee, J.; Wlodkowski, A.; Karunakaran, E.; Biggs, C.A.; Wright, P.C.; Albers, S.V. Macromolecular fingerprinting of Sulfolobus species in biofilm: A transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 2011, 10, 4105–4119. [Google Scholar] [CrossRef]
- Van Wolferen, M.; Orell, A.; Albers, S.V. Archaeal biofilm formation. Nat. Rev. Microbiol. 2018, 16, 699–713. [Google Scholar] [CrossRef]
- Braun, F.; Recalde, A.; Bähre, H.; Seifert, R.; Albers, S.V. Putative nucleotide-based second messengers in the archaeal model organisms Haloferax volcanii and Sulfolobus acidocaldarius. Front. Microbiol. 2021, 12, 779012. [Google Scholar] [CrossRef]
- Orell, A.; Peeters, E.; Vassen, V.; Jachlewski, S.; Schalles, S.; Siebers, B.; Albers, S.V. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 2013, 768, 1886–1898. [Google Scholar] [CrossRef]
- Li, L.; Banerjee, A.; Bischof, L.F.; Maklad, H.R.; Hoffmann, L.; Henche, A.L.; Veliz, F.; Bildl, W.; Schulte, U.; Orell, A.; et al. Wing phosphorylation is a major functional determinant of the Lrs14-type biofilm and motility regulator AbfR1 in Sulfolobus acidocaldarius. Mol. Microbiol. 2017, 105, 777–793. [Google Scholar] [CrossRef]
- Lewis, A.M.; Willard, D.J.; HManesh, M.J.; Sivabalasarma, S.; Albers, S.V.; Kelly, R.M. Stay or go: Sulfolobales biofilm dispersal is dependent on a bifunctional VapB antitoxin. Mbio 2023, 14, e00053-23. [Google Scholar] [CrossRef]
- Recalde, A.; van Wolferen, M.; Sivabalasarma, S.; Albers, S.V.; Navarro, C.A.; Jerez, C.A. The role of polyphosphate in motility, adhesion, and biofilm formation in Sulfolobales. Microorganisms 2021, 9, 193. [Google Scholar] [CrossRef]
- Laplagia, C.; Hartzell, P.L. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 1997, 63, 3158–3163. [Google Scholar] [CrossRef]
- Koechler, S.; Farasin, J.; Cleiss-Arnold, J.; Arsène-Ploetze, F. Toxic metal resistance in biofilms: Diversity of microbial responses and their evolution. Res. Microbiol. 2015, 166, 764–773. [Google Scholar] [CrossRef]
- Zhang, R.; Neu, T.R.; Zhang, Y.; Bellenberg, S.; Kuhlicke, U.; Li, Q.; Sand, W.; Vera, M. Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl. Microbiol. Biotechnol. 2015, 99, 7343–7356. [Google Scholar] [CrossRef]
- Castro, C.; Zhang, R.; Liu, J.; Bellenberg, S.; Neu, T.R.; Donati, E.; Sand, W.; Vera, M. Biofilm formation and interspecies interactions in mixed cultures of thermo-acidophilic archaea Acidianus spp. and Sulfolobus metallicus. Res. Microbiol. 2016, 167, 604–612. [Google Scholar] [CrossRef]
- Safar, C.; Castro, C.; Donati, E. Importance of initial interfacial steps during chalcopyrite bioleaching by a thermoacidophilic archaeon. Microorganisms 2020, 8, 1009. [Google Scholar] [CrossRef]
- Martínez-Bussenius, C.; Navarro, C.A.; Jerez, C.A. Microbial copper resistance: Importance in biohydrometallurgy. Microb. Biotechnol. 2017, 10, 279–295. [Google Scholar] [CrossRef]
- McCarthy, S.; Ai, C.; Wheaton, G.; Tevatia, R.; Eckrich, V.; Kelly, R.; Blum, P. Role of an Archaeal PitA Transporter in the Copper and Arsenic Resistance of Metallosphaera sedula, an Extreme Thermoacidophile. J. Bacteriol. 2014, 196, 3562–3570. [Google Scholar] [CrossRef]
- Orell, A.; Remonsellez, F.; Arancibia, R.; Jerez, C.A. Molecular characterization of copper and cadmium resistance determinants in the biomining thermoacidophilic archaeon Sulfolobus metallicus. Archaea 2013, 2013, 289236. [Google Scholar] [CrossRef]
- Soto, D.F.; Recalde, A.; Orell, A.; Albers, S.V.; Paradela, A.; Navarro, C.A.; Jerez, C.A. Global effect of the lack of inorganic polyphosphate in the extremophilic archaeon Sulfolobus solfataricus: A proteomic approach. J. Proteomics 2019, 191, 143–152. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Zolghadr, B.; Klingl, A.; Koerdt, A.; Driessen, A.J.; Rachel, R.; Albers, S.V. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 2010, 192, 104–110. [Google Scholar] [CrossRef]
- Koerdt, A.; Jachlewski, S.; Ghosh, A.; Wingender, J.; Siebers, B.; Albers, S.V. Complementation of Sulfolobus solfataricus PBL2025 with an α-mannosidase: Effects on surface attachment and biofilm formation. Extremophiles 2012, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Völlmecke, C.; Drees, S.L.; Reimann, J.; Albers, S.V.; Lübben, M. The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 2012, 158 Pt 6, 1622–1633. [Google Scholar] [CrossRef]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Eichler, J. Extreme sweetness: Protein glycosylation in archaea. Nat. Rev. Microbiol. 2013, 11, 151–156. [Google Scholar] [CrossRef]
- Guan, Z.; Naparstek, S.; Calo, D.; Eichler, J. Protein glycosylation as an adaptive response in Archaea: Growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ. Microbiol. 2012, 14, 743–753. [Google Scholar] [CrossRef]
- Huo, X.; Yang, H.; Feng, S. Insights into the defensive mechanism of bioleaching microorganisms under extreme environmental copper stress. Rev. Environ. Sci. Biotechnol. 2023, 22, 79–103. [Google Scholar] [CrossRef]
- Valdebenito-Rolack, E.; Ruiz-Tagle, N.; Abarzúa, L.; Aroca, G.; Urrutia, H. Characterization of a hyperthermophilic sulphur-oxidizing biofilm produced by archaea isolated from a hot spring. Electron. J. Biotechnol. 2017, 25, 58–63. [Google Scholar] [CrossRef]
- Liu, L.Z.; Nie, Z.Y.; Yang, Y.; Pan, X.; Xia, X.; Zhou, Y.H.; Xia, J.L.; Zhang, L.J.; Zhen, X.J.; Yang, H.Y. In situ characterization of change in superficial organic components of thermoacidophilic archaeon Acidianus manzaensis YN-25. Res. Microbiol. 2018, 169, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Neu, T.R.; Blanchard, V.; Vera, M.; Sand, W. Biofilm dynamics and EPS production of a thermoacidophilic bioleaching archaeon. New Biotechnol. 2019, 51, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Villafane, A.; Voskoboynik, Y.; Cuebas, M.; Ruhl, I.; Bini, E. Response to excess copper in the hyperthermophile Sulfolobus solfataricus strain 98/2. Biochem. Biophys. Res. Commun. 2010, 385, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Benninghoff, J.C.; Kuschmierz, L.; Zhou, X.; Albersmeier, A.; Pham, T.K.; Busche, T.; Wright, P.C.; Kalinowski, J.; Makarova, K.S.; Bräsen, C.; et al. Exposure to 1-butanol exemplifies the response of the thermoacidophilic Archaeon Sulfolobus acidocaldarius to solvent stress. Appl. Environ. Microbiol. 2021, 87, e02988-20. [Google Scholar] [CrossRef]
- Meyer, B.H.; Zolghadr, B.; Peyfoon, E.; Pabst, M.; Panico, M.; Morris, H.R.; Haslan, S.M.; Messner, P.; Schäffer, C.; Dell, A.; et al. Sulfoquinovose synthase–an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol. Microbiol. 2011, 82, 1150–1163. [Google Scholar] [CrossRef]
- Meyer, B.H.; Peyfoon, E.; Dietrich, C.; Hitchen, P.; Panico, M.; Morris, H.R.; Dell, A.; Albers, S.V. Agl16, a thermophilic glycosyltransferase mediating the last step of N-glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bacteriol. 2013, 195, 2177–2186. [Google Scholar] [CrossRef]
- Schopf, S.; Wanner, G.; Rachel, R.; Wirth, R. An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri. Arch. Microbiol. 2008, 190, 371–377. [Google Scholar] [CrossRef]
- Vargas-Straube, M.J.; Beard, S.; Norambuena, R.; Paradela, A.; Vera, M.; Jerez, C.A. High copper concentration reduces biofilm formation in Acidithiobacillus ferrooxidans by decreasing production of extracellular polymeric substances and its adherence to elemental sulfur. J. Proteomics. 2020, 225, 103874. [Google Scholar] [CrossRef]
- Vega, L.M.; Mathieu, J.; Yang, Y.; Pyle, B.H.; McLean, R.J.; Alvarez, P.J. Nickel and cadmium ions inhibit quorum sensing and biofilm formation without affecting viability in Burkholderia multivorans. Int. Biodeterior. Biodegrad. 2014, 91, 82–87. [Google Scholar] [CrossRef]
- Albers, S.V.; Jarrell, K.F. The archaellum: How Archaea swim. Front. Microbiol. 2015, 6, 23. [Google Scholar] [CrossRef]
- Feng, S.; Hou, S.; Cui, Y.; Tong, Y.; Yang, H. Metabolic transcriptional analysis on copper tolerance in moderate thermophilic bioleaching microorganism Acidithiobacillus caldus. J. Ind. Microbiol. Biotechnol. 2020, 47, 21–33. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recalde, A.; González-Madrid, G.; Acevedo-López, J.; Jerez, C.A. Sessile Lifestyle Offers Protection against Copper Stress in Saccharolobus solfataricus. Microorganisms 2023, 11, 1421. https://doi.org/10.3390/microorganisms11061421
Recalde A, González-Madrid G, Acevedo-López J, Jerez CA. Sessile Lifestyle Offers Protection against Copper Stress in Saccharolobus solfataricus. Microorganisms. 2023; 11(6):1421. https://doi.org/10.3390/microorganisms11061421
Chicago/Turabian StyleRecalde, Alejandra, Gabriela González-Madrid, José Acevedo-López, and Carlos A. Jerez. 2023. "Sessile Lifestyle Offers Protection against Copper Stress in Saccharolobus solfataricus" Microorganisms 11, no. 6: 1421. https://doi.org/10.3390/microorganisms11061421






